Insights into the Molecular Basis of Huanglongbing Tolerance in Persian Lime (Citrus latifolia Tan.) through a Transcriptomic Approach
- PMID: 37108662
- PMCID: PMC10144405
- DOI: 10.3390/ijms24087497
Insights into the Molecular Basis of Huanglongbing Tolerance in Persian Lime (Citrus latifolia Tan.) through a Transcriptomic Approach
Abstract
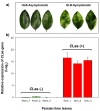
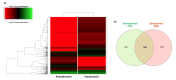
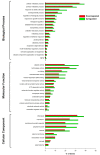
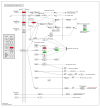
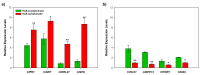
Huanglongbing (HLB) is a vascular disease of Citrus caused by three species of the α-proteobacteria "Candidatus Liberibacter", with "Candidatus Liberibacter asiaticus" (CLas) being the most widespread and the one causing significant economic losses in citrus-producing regions worldwide. However, Persian lime (Citrus latifolia Tanaka) has shown tolerance to the disease. To understand the molecular mechanisms of this tolerance, transcriptomic analysis of HLB was performed using asymptomatic and symptomatic leaves. RNA-Seq analysis revealed 652 differentially expressed genes (DEGs) in response to CLas infection, of which 457 were upregulated and 195 were downregulated. KEGG analysis revealed that after CLas infection, some DEGs were present in the plant-pathogen interaction and in the starch and sucrose metabolism pathways. DEGs present in the plant-pathogen interaction pathway suggests that tolerance against HLB in Persian lime could be mediated, at least partly, by the ClRSP2 and ClHSP90 genes. Previous reports documented that RSP2 and HSP90 showed low expression in susceptible citrus genotypes. Regarding the starch and sucrose metabolism pathways, some genes were identified as being related to the imbalance of starch accumulation. On the other hand, eight biotic stress-related genes were selected for further RT-qPCR analysis to validate our results. RT-qPCR results confirmed that symptomatic HLB leaves had high relative expression levels of the ClPR1, ClNFP, ClDR27, and ClSRK genes, whereas the ClHSL1, ClRPP13, ClPDR1, and ClNAC genes were expressed at lower levels than those from HLB asymptomatic leaves. Taken together, the present transcriptomic analysis contributes to the understanding of the CLas-Persian lime interaction in its natural environment and may set the basis for developing strategies for the integrated management of this important Citrus disease through the identification of blanks for genetic improvement.
Keywords: Huanglongbing; basal resistance; citrus; pathogen triggered immunity; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Microscopic and Transcriptomic Analyses of Early Events Triggered by 'Candidatus Liberibacter asiaticus' in Young Flushes of Huanglongbing-Positive Citrus Trees.Phytopathology. 2023 Jun;113(6):985-997. doi: 10.1094/PHYTO-10-22-0360-R. Epub 2023 Aug 2. Phytopathology. 2023. PMID: 36449527
-
Comprehensive Transcriptomic Profiling of Citrus australasica Unveils Antimicrobial Peptides and Immune Pathways for Huanglongbing Tolerance.J Agric Food Chem. 2025 Jul 9;73(27):16847-16859. doi: 10.1021/acs.jafc.4c11910. Epub 2025 Jun 11. J Agric Food Chem. 2025. PMID: 40497554
-
Transcriptomic and biochemical analysis of pummelo x finger lime hybrids in response to Huanglongbing (HLB).BMC Plant Biol. 2025 Feb 20;25(1):235. doi: 10.1186/s12870-025-06211-8. BMC Plant Biol. 2025. PMID: 39979795 Free PMC article.
-
Molecular signatures between citrus and Candidatus Liberibacter asiaticus.PLoS Pathog. 2021 Dec 9;17(12):e1010071. doi: 10.1371/journal.ppat.1010071. eCollection 2021 Dec. PLoS Pathog. 2021. PMID: 34882744 Free PMC article. Review.
-
Progress and Obstacles in Culturing 'Candidatus Liberibacter asiaticus', the Bacterium Associated with Huanglongbing.Phytopathology. 2019 Jul;109(7):1092-1101. doi: 10.1094/PHYTO-02-19-0051-RVW. Epub 2019 Jun 3. Phytopathology. 2019. PMID: 30998129 Review.
Cited by
-
Recent progress in the understanding of Citrus Huanglongbing: from the perspective of pathogen and citrus host.Mol Breed. 2024 Nov 6;44(11):77. doi: 10.1007/s11032-024-01517-1. eCollection 2024 Nov. Mol Breed. 2024. PMID: 39525404
References
-
- Méndez J.M.A., Caamal V.J.H., Rodríguez A.N.L., Vargas D.A.A., Alamilla M.J.C., Criollo C.M.A. Avances y limitantes en la micropropagación del limón persa (Citrus × latifolia Tan.) Temas Cienc. Tecnol. 2020;34:33–40.
-
- SADER. Secretaría de Agricultura y Desarrollo Rural Planeación Agrícola Citrícola Nacional. [(accessed on 1 December 2022)];2021 Available online: www.gob.mx/cms/uploads/attachment/file/257073/Potencial-C_tricos.
-
- SIAP. Servicio de Información Agroalimentaria y Pesquera Datos Abiertos de México. Producción Anual Agrícola de Cítricos en México. [(accessed on 1 December 2022)];2020 Available online: https://datos.gob.mx/busca/organization/siap.
-
- Berdeja-Arbeu R., Gómez M., Méndez-Gómez J., Escobar-Hernández R., Pérez-Marroquín G. Rendimiento y calidad de fruta de lima ‘Persa’ con nutrición química, estiércol y leguminosa en Martínez de la Torre, Veracruz, México. Investig. Cienc. Univ. Autónoma Aguascalientes. 2019;1:44–50. doi: 10.33064/iycuaa2019782231. - DOI
-
- Navarro L. Microinjerto de Ápices Caulinares In Vitro para la Obtención de Plantas de Agrios Libres de Virus. [(accessed on 1 December 2022)];Bol. Serv. Plagas. 1979 5:127–148. Available online: https://redivia.gva.es/handle/20.500.11939/7363.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

