Comparative Genomic Analysis of a Methylorubrum rhodesianum MB200 Isolated from Biogas Digesters Provided New Insights into the Carbon Metabolism of Methylotrophic Bacteria
- PMID: 37108681
- PMCID: PMC10138955
- DOI: 10.3390/ijms24087521
Comparative Genomic Analysis of a Methylorubrum rhodesianum MB200 Isolated from Biogas Digesters Provided New Insights into the Carbon Metabolism of Methylotrophic Bacteria
Abstract
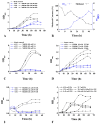
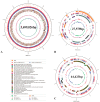
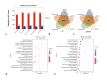
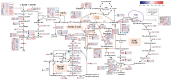
Methylotrophic bacteria are widely distributed in nature and can be applied in bioconversion because of their ability to use one-carbon source. The aim of this study was to investigate the mechanism underlying utilization of high methanol content and other carbon sources by Methylorubrum rhodesianum strain MB200 via comparative genomics and analysis of carbon metabolism pathway. The genomic analysis revealed that the strain MB200 had a genome size of 5.7 Mb and two plasmids. Its genome was presented and compared with that of the 25 fully sequenced strains of Methylobacterium genus. Comparative genomics revealed that the Methylorubrum strains had closer collinearity, more shared orthogroups, and more conservative MDH cluster. The transcriptome analysis of the strain MB200 in the presence of various carbon sources revealed that a battery of genes was involved in the methanol metabolism. These genes are involved in the following functions: carbon fixation, electron transfer chain, ATP energy release, and resistance to oxidation. Particularly, the central carbon metabolism pathway of the strain MB200 was reconstructed to reflect the possible reality of the carbon metabolism, including ethanol metabolism. Partial propionate metabolism involved in ethyl malonyl-CoA (EMC) pathway might help to relieve the restriction of the serine cycle. In addition, the glycine cleavage system (GCS) was observed to participate in the central carbon metabolism pathway. The study revealed the coordination of several metabolic pathways, where various carbon sources could induce associated metabolic pathways. To the best of our knowledge, this is the first study providing a more comprehensive understanding of the central carbon metabolism in Methylorubrum. This study provided a reference for potential synthetic and industrial applications of this genus and its use as chassis cells.
Keywords: Methylorubrum rhodesianum MB200; carbon metabolism; comparative genomics; methanol.
Conflict of interest statement
We declare no conflicts of interest.
Figures

References
-
- Green P.N. Methylobacterium Patt, Cole and Hanson 1976, 228AL emend. Green and Bousfield 1983, 876. In: Brenner D.J., Krieg N.R., Garrity G.M., Staley J.T., Boone D.R., Vos P., Goodfellow M., Rainey F.A., Schleifer K.-H., editors. Bergey’s Manual® of Systematic Bacteriology. Springer; New York, NY, USA: 2005. pp. 567–571. - DOI
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

