Multiple Nucleocapsid Structural Forms of Shrimp White Spot Syndrome Virus Suggests a Novel Viral Morphogenetic Pathway
- PMID: 37108688
- PMCID: PMC10140842
- DOI: 10.3390/ijms24087525
Multiple Nucleocapsid Structural Forms of Shrimp White Spot Syndrome Virus Suggests a Novel Viral Morphogenetic Pathway
Abstract
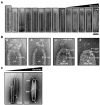
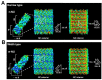
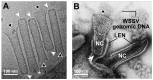
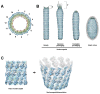
White spot syndrome virus (WSSV) is a very large dsDNA virus. The accepted shape of the WSSV virion has been as ellipsoidal, with a tail-like extension. However, due to the scarcity of reliable references, the pathogenesis and morphogenesis of WSSV are not well understood. Here, we used transmission electron microscopy (TEM) and cryogenic electron microscopy (Cryo-EM) to address some knowledge gaps. We concluded that mature WSSV virions with a stout oval-like shape do not have tail-like extensions. Furthermore, there were two distinct ends in WSSV nucleocapsids: a portal cap and a closed base. A C14 symmetric structure of the WSSV nucleocapsid was also proposed, according to our Cryo-EM map. Immunoelectron microscopy (IEM) revealed that VP664 proteins, the main components of the 14 assembly units, form a ring-like architecture. Moreover, WSSV nucleocapsids were also observed to undergo unique helical dissociation. Based on these new results, we propose a novel morphogenetic pathway of WSSV.
Keywords: cryogenic electron microscopy; large dsDNA virus; nucleocapsid; white spot syndrome virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Identification of the nucleocapsid, tegument, and envelope proteins of the shrimp white spot syndrome virus virion.J Virol. 2006 Mar;80(6):3021-9. doi: 10.1128/JVI.80.6.3021-3029.2006. J Virol. 2006. PMID: 16501111 Free PMC article.
-
The unique stacked rings in the nucleocapsid of the white spot syndrome virus virion are formed by the major structural protein VP664, the largest viral structural protein ever found.J Virol. 2005 Jan;79(1):140-9. doi: 10.1128/JVI.79.1.140-149.2005. J Virol. 2005. PMID: 15596810 Free PMC article.
-
Shotgun identification of the structural proteome of shrimp white spot syndrome virus and iTRAQ differentiation of envelope and nucleocapsid subproteomes.Mol Cell Proteomics. 2007 Sep;6(9):1609-20. doi: 10.1074/mcp.M600327-MCP200. Epub 2007 Jun 2. Mol Cell Proteomics. 2007. PMID: 17545682
-
A review on the morphology, molecular characterization, morphogenesis and pathogenesis of white spot syndrome virus.J Fish Dis. 2008 Jan;31(1):1-18. doi: 10.1111/j.1365-2761.2007.00877.x. J Fish Dis. 2008. PMID: 18086030 Review.
-
A model for apoptotic interaction between white spot syndrome virus and shrimp.Fish Shellfish Immunol. 2013 Apr;34(4):1011-7. doi: 10.1016/j.fsi.2012.05.030. Epub 2012 Jun 7. Fish Shellfish Immunol. 2013. PMID: 22683516 Review.
Cited by
-
The nucleocapsid architecture and structural atlas of the prototype baculovirus define the hallmarks of a new viral realm.Sci Adv. 2024 Dec 20;10(51):eado2631. doi: 10.1126/sciadv.ado2631. Epub 2024 Dec 18. Sci Adv. 2024. PMID: 39693434 Free PMC article.
References
-
- Dey B.K., Dugassa G.H., Hinzano S.M., Bossier P. Causative agent, diagnosis and management of white spot disease in shrimp: A review. Rev. Aquac. 2020;12:822–865. doi: 10.1111/raq.12352. - DOI
-
- Chou H.Y., Huang C.Y., Wang C.H., Chiang H.C., Lo C.F. Pathogenicity of a baculovirus infection causing White Spot Syndrome in cultured Penaeid shrimp in Taiwan. Dis. Aquat. Organ. 1995;23:165–173. doi: 10.3354/dao023165. - DOI
MeSH terms
Grants and funding
- MOST 108-2314-B-006-096-MY3; MOST 108-2112-M-002-016-MY3; MOST 109-2634-F-006-022; MOST 110-2313-B-019-004-MY3/Ministry of Science and Technology
- NSTC 111-2628-B-006-017/National Science and Technology Council
- MOE 106R880708/Ministry of Education
- AS-KPQ-109-TPP2; AS-KPQ-105-TPP/Taiwan Protein Project
- AS-CFII-111-210/Academia Sinica Cryo-EM Center
LinkOut - more resources
Full Text Sources
Miscellaneous

