Mitoregulin Contributes to Creatine Shuttling and Cardiolipin Protection in Mice Muscle
- PMID: 37108753
- PMCID: PMC10143810
- DOI: 10.3390/ijms24087589
Mitoregulin Contributes to Creatine Shuttling and Cardiolipin Protection in Mice Muscle
Abstract
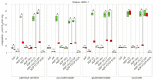
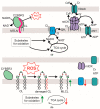
Small peptides compose a large share of the mitochondrial proteome. Mitoregulin (Mtln) is a mitochondrial peptide known to contribute to the respiratory complex I functioning and other processes in mitochondria. In our previous studies, we demonstrated that Mtln knockout mice develop obesity and accumulate triglycerides and other oxidation substrates in serum, concomitant with an exhaustion of tricarboxylic acids cycle intermediates. Here we examined the functional role of Mtln in skeletal muscles, one of the major energy consuming tissues. We observed reduced muscle strength for Mtln knockout mice. Decrease of the mitochondrial cardiolipin and concomitant increase in monolysocardiolipin concentration upon Mtln inactivation is likely to be a consequence of imbalance between oxidative damage and remodeling of cardiolipin. It is accompanied by the mitochondrial creatine kinase octamer dissociation and suboptimal respiratory chain performance in Mtln knockout mice.
Keywords: cardiolipin; creatine kinase; metabolism; mitochondria; oxidative phosphorylation; short open reading frame; small peptide.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Kidney-Related Function of Mitochondrial Protein Mitoregulin.Int J Mol Sci. 2023 May 22;24(10):9106. doi: 10.3390/ijms24109106. Int J Mol Sci. 2023. PMID: 37240452 Free PMC article.
-
Mitoregulin, a tiny protein at the crossroads of mitochondrial functioning, stress, and disease.Front Cell Dev Biol. 2025 Mar 5;13:1545359. doi: 10.3389/fcell.2025.1545359. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40109364 Free PMC article. Review.
-
Mitochondrial peptide Mtln contributes to oxidative metabolism in mice.Biochimie. 2023 Jan;204:136-139. doi: 10.1016/j.biochi.2022.09.009. Epub 2022 Sep 26. Biochimie. 2023. PMID: 36174793
-
Mitoregulin: A lncRNA-Encoded Microprotein that Supports Mitochondrial Supercomplexes and Respiratory Efficiency.Cell Rep. 2018 Jun 26;23(13):3710-3720.e8. doi: 10.1016/j.celrep.2018.06.002. Cell Rep. 2018. PMID: 29949756 Free PMC article.
-
Role of cardiolipin in skeletal muscle function and its therapeutic implications.Cell Commun Signal. 2025 Jan 21;23(1):36. doi: 10.1186/s12964-025-02032-2. Cell Commun Signal. 2025. PMID: 39833875 Free PMC article. Review.
Cited by
-
Non-canonical ORFs-derived protein products in mitochondria: A multifaceted exploration of their functions in health and disease.Protein Sci. 2025 Mar;34(3):e70053. doi: 10.1002/pro.70053. Protein Sci. 2025. PMID: 39969119 Free PMC article. Review.
-
Mitoregulin Promotes Cell Cycle Progression in Non-Small Cell Lung Cancer Cells.Int J Mol Sci. 2025 Feb 24;26(5):1939. doi: 10.3390/ijms26051939. Int J Mol Sci. 2025. PMID: 40076565 Free PMC article.
-
Kidney-Related Function of Mitochondrial Protein Mitoregulin.Int J Mol Sci. 2023 May 22;24(10):9106. doi: 10.3390/ijms24109106. Int J Mol Sci. 2023. PMID: 37240452 Free PMC article.
-
Mitoregulin, a tiny protein at the crossroads of mitochondrial functioning, stress, and disease.Front Cell Dev Biol. 2025 Mar 5;13:1545359. doi: 10.3389/fcell.2025.1545359. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40109364 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

