Machine Learning Identifies New Predictors on Restenosis Risk after Coronary Artery Stenting in 10,004 Patients with Surveillance Angiography
- PMID: 37109283
- PMCID: PMC10142067
- DOI: 10.3390/jcm12082941
Machine Learning Identifies New Predictors on Restenosis Risk after Coronary Artery Stenting in 10,004 Patients with Surveillance Angiography
Abstract
Objective: Machine learning (ML) approaches have the potential to uncover regular patterns in multi-layered data. Here we applied self-organizing maps (SOMs) to detect such patterns with the aim to better predict in-stent restenosis (ISR) at surveillance angiography 6 to 8 months after percutaneous coronary intervention with stenting.
Methods: In prospectively collected data from 10,004 patients receiving percutaneous coronary intervention (PCI) for 15,004 lesions, we applied SOMs to predict ISR angiographically 6-8 months after index procedure. SOM findings were compared with results of conventional uni- and multivariate analyses. The predictive value of both approaches was assessed after random splitting of patients into training and test sets (50:50).
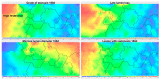
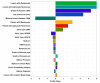
Results: Conventional multivariate analyses revealed 10, mostly known, predictors for restenosis after coronary stenting: balloon-to-vessel ratio, complex lesion morphology, diabetes mellitus, left main stenting, stent type (bare metal vs. first vs. second generation drug eluting stent), stent length, stenosis severity, vessel size reduction, and prior bypass surgery. The SOM approach identified all these and nine further predictors, including chronic vessel occlusion, lesion length, and prior PCI. Moreover, the SOM-based model performed well in predicting ISR (AUC under ROC: 0.728); however, there was no meaningful advantage in predicting ISR at surveillance angiography in comparison with the conventional multivariable model (0.726, p = 0.3).
Conclusions: The agnostic SOM-based approach identified-without clinical knowledge-even more contributors to restenosis risk. In fact, SOMs applied to a large prospectively sampled cohort identified several novel predictors of restenosis after PCI. However, as compared with established covariates, ML technologies did not improve identification of patients at high risk for restenosis after PCI in a clinically relevant fashion.
Keywords: artificial intelligence; coronary artery disease; machine learning; percutaneous coronary intervention; prediction; restenosis.
Conflict of interest statement
B.G and D.M. are affiliated to Biomax Informatics AG; H.S. reports honoraria from AstraZeneca, Bayer Vital GmbH, MSD Sharp & Dohme, Novartis, Servier, Sanofi-Aventis, Brahms, Brystol-Myers Sqibb, Medtronic, Boehringer Ingelheim, Daiichi Sankyo, AMGEN, Pfizer, Synlab and consulting fees from AstraZeneca, Amgen, MSD Sharp & Dohme, not related to the current work; All other authors declare no conflict of interest.
Figures

References
-
- Fortmeier V., Lachmann M., Korber M.I., Unterhuber M., von Scheidt M., Rippen E., Harmsen G., Gercek M., Friedrichs K.P., Roder F., et al. Solving the Pulmonary Hypertension Paradox in Patients With Severe Tricuspid Regurgitation by Employing Artificial Intelligence. JACC Cardiovasc. Interv. 2022;15:381–394. doi: 10.1016/j.jcin.2021.12.043. - DOI - PubMed
-
- Fortmeier V., Lachmann M., Unterhuber M., Stolz L., Kassar M., Ochs L., Gercek M., Schober A.R., Stocker T.J., Omran H., et al. Epiphenomenon or Prognostically Relevant Interventional Target? A Novel Proportionality Framework for Severe Tricuspid Regurgitation. J. Am. Heart Assoc. 2023;12:e028737. doi: 10.1161/JAHA.122.028737. - DOI - PMC - PubMed
-
- Lachmann M., Rippen E., Rueckert D., Schuster T., Xhepa E., von Scheidt M., Pellegrini C., Trenkwalder T., Rheude T., Stundl A., et al. Harnessing feature extraction capacities from a pre-trained convolutional neural network (VGG-16) for the unsupervised distinction of aortic outflow velocity profiles in patients with severe aortic stenosis. Eur. Heart J. Digit. Health. 2022;3:153–168. doi: 10.1093/ehjdh/ztac004. - DOI - PMC - PubMed
Grants and funding
- ZF4590201BA8/German Federal Ministry of Economics and Energy
- 01ZX1904A/German Federal Ministry of Education and Research
- DZHK-BHF: 81X2600522/British Heart Foundation (BHF)/German Centre of Cardiovascular Research (DZHK)-collaboration
- PlaqOmics: 18CVD02/Leducq Foundation for Cardiovascular Research
- DMB-1805-0001/Bavarian State Ministry of Health and Care
LinkOut - more resources
Full Text Sources
Miscellaneous

