Structural Analysis of Bacillus subtilis Sigma Factors
- PMID: 37110501
- PMCID: PMC10141391
- DOI: 10.3390/microorganisms11041077
Structural Analysis of Bacillus subtilis Sigma Factors
Abstract
Bacteria use an array of sigma factors to regulate gene expression during different stages of their life cycles. Full-length, atomic-level structures of sigma factors have been challenging to obtain experimentally as a result of their many regions of intrinsic disorder. AlphaFold has now supplied plausible full-length models for most sigma factors. Here we discuss the current understanding of the structures and functions of sigma factors in the model organism, Bacillus subtilis, and present an X-ray crystal structure of a region of B. subtilis SigE, a sigma factor that plays a critical role in the developmental process of spore formation.
Keywords: Alphafold; Bacillus subtilis; SigE; X-ray crystallography; sigma factor; sporulation; structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
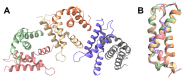


References
Grants and funding
LinkOut - more resources
Full Text Sources

