Computational Insights into the Dynamic Structural Features and Binding Characteristics of Recombinase UvsX Compared with RecA
- PMID: 37110596
- PMCID: PMC10144138
- DOI: 10.3390/molecules28083363
Computational Insights into the Dynamic Structural Features and Binding Characteristics of Recombinase UvsX Compared with RecA
Abstract
RecA family recombinases are the core enzymes in the process of homologous recombination, and their normal operation ensures the stability of the genome and the healthy development of organisms. The UvsX protein from bacteriophage T4 is a member of the RecA family recombinases and plays a central role in T4 phage DNA repair and replication, which provides an important model for the biochemistry and genetics of DNA metabolism. UvsX shares a high degree of structural similarity and function with RecA, which is the most deeply studied member of the RecA family. However, the detailed molecular mechanism of UvsX has not been resolved. In this study, a comprehensive all-atom molecular dynamics simulation of the UvsX protein dimer complex was carried out in order to investigate the conformational and binding properties of UvsX in combination with ATP and DNA, and the simulation of RecA was synchronized with the property comparison learning for UvsX. This study confirmed the highly conserved molecular structure characteristics and catalytic centers of RecA and UvsX, and also discovered differences in regional conformation, volatility and the ability to bind DNA between the two proteins at different temperatures, which would be helpful for the subsequent understanding and application of related recombinases.
Keywords: DNA recombinases; RecA; UvsX; homologous modelling; molecular dynamics simulations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



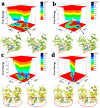

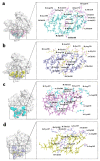
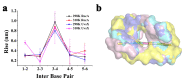

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

