A Retrospective Assessment of Sputum Samples and Antimicrobial Resistance in COVID-19 Patients
- PMID: 37111506
- PMCID: PMC10143659
- DOI: 10.3390/pathogens12040620
A Retrospective Assessment of Sputum Samples and Antimicrobial Resistance in COVID-19 Patients
Abstract
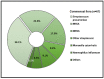
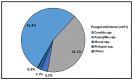
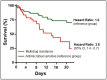
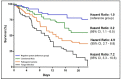
Data on bacterial or fungal pathogens and their impact on the mortality rates of Western Romanian COVID-19 patients are scarce. As a result, the purpose of this research was to determine the prevalence of bacterial and fungal co- and superinfections in Western Romanian adults with COVID-19, hospitalized in in-ward settings during the second half of the pandemic, and its distribution according to sociodemographic and clinical conditions. The unicentric retrospective observational study was conducted on 407 eligible patients. Expectorate sputum was selected as the sampling technique followed by routine microbiological investigations. A total of 31.5% of samples tested positive for Pseudomonas aeruginosa, followed by 26.2% having co-infections with Klebsiella pneumoniae among patients admitted with COVID-19. The third most common Pathogenic bacteria identified in the sputum samples was Escherichia coli, followed by Acinetobacter baumannii in 9.3% of samples. Commensal human pathogens caused respiratory infections in 67 patients, the most prevalent being Streptococcus penumoniae, followed by methicillin-sensitive and methicillin-resistant Staphylococcus aureus. A total of 53.4% of sputum samples tested positive for Candida spp., followed by 41.1% of samples with Aspergillus spp. growth. The three groups with positive microbial growth on sputum cultures had an equally proportional distribution of patients admitted to the ICU, with an average of 30%, compared with only 17.3% among hospitalized COVID-19 patients with negative sputum cultures (p = 0.003). More than 80% of all positive samples showed multidrug resistance. The high prevalence of bacterial and fungal co-infections and superinfections in COVID-19 patients mandates for strict and effective antimicrobial stewardship and infection control policies.
Keywords: SARS-CoV-2; co-infection; multidrug resistance; outcome; superinfection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Kariyawasam R.M., Julien D.A., Jelinski D.C., Larose S.L., Rennert-May E., Conly J.M., Dingle T.C., Chen J.Z., Tyrrell G.J., Ronksley P.E., et al. Antimicrobial Resistance (AMR) in COVID-19 Patients: A Systematic Review and Meta-Analysis (November 2019–June 2021) Antimicrob. Resist. Infect. Control. 2022;11:45. doi: 10.1186/s13756-022-01085-z. - DOI - PMC - PubMed
-
- Rotondo J.C., Martini F., Maritati M., Caselli E., Gallenga C.E., Guarino M., De Giorgio R., Mazziotta C., Tramarin M.L., Badiale G., et al. Advanced Molecular and Immunological Diagnostic Methods to Detect SARS-CoV-2 Infection. Microorganisms. 2022;10:1193. doi: 10.3390/microorganisms10061193. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

