Development and Validation of an Explainable Machine Learning-Based Prediction Model for Drug-Food Interactions from Chemical Structures
- PMID: 37112302
- PMCID: PMC10143839
- DOI: 10.3390/s23083962
Development and Validation of an Explainable Machine Learning-Based Prediction Model for Drug-Food Interactions from Chemical Structures
Abstract
Possible drug-food constituent interactions (DFIs) could change the intended efficiency of particular therapeutics in medical practice. The increasing number of multiple-drug prescriptions leads to the rise of drug-drug interactions (DDIs) and DFIs. These adverse interactions lead to other implications, e.g., the decline in medicament's effect, the withdrawals of various medications, and harmful impacts on the patients' health. However, the importance of DFIs remains underestimated, as the number of studies on these topics is constrained. Recently, scientists have applied artificial intelligence-based models to study DFIs. However, there were still some limitations in data mining, input, and detailed annotations. This study proposed a novel prediction model to address the limitations of previous studies. In detail, we extracted 70,477 food compounds from the FooDB database and 13,580 drugs from the DrugBank database. We extracted 3780 features from each drug-food compound pair. The optimal model was eXtreme Gradient Boosting (XGBoost). We also validated the performance of our model on one external test set from a previous study which contained 1922 DFIs. Finally, we applied our model to recommend whether a drug should or should not be taken with some food compounds based on their interactions. The model can provide highly accurate and clinically relevant recommendations, especially for DFIs that may cause severe adverse events and even death. Our proposed model can contribute to developing more robust predictive models to help patients, under the supervision and consultants of physicians, avoid DFI adverse effects in combining drugs and foods for therapy.
Keywords: DrugBank; FooDB; adverse food reaction; chemical informatics; drug–food interactions; drug–nutrient interactions; explainable artificial intelligence; machine learning; precision medicine; simplified molecular-input line-entry system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

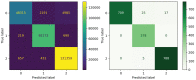
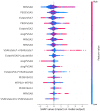
Similar articles
-
Update and Application of a Deep Learning Model for the Prediction of Interactions between Drugs Used by Patients with Multiple Sclerosis.Pharmaceutics. 2023 Dec 19;16(1):3. doi: 10.3390/pharmaceutics16010003. Pharmaceutics. 2023. PMID: 38276481 Free PMC article.
-
An AI-based Prediction Model for Drug-drug Interactions in Osteoporosis and Paget's Diseases from SMILES.Mol Inform. 2022 Jun;41(6):e2100264. doi: 10.1002/minf.202100264. Epub 2022 Jan 22. Mol Inform. 2022. PMID: 34989149
-
Explainable Machine Learning Techniques To Predict Amiodarone-Induced Thyroid Dysfunction Risk: Multicenter, Retrospective Study With External Validation.J Med Internet Res. 2023 Feb 7;25:e43734. doi: 10.2196/43734. J Med Internet Res. 2023. PMID: 36749620 Free PMC article.
-
Computational and artificial intelligence-based approaches for drug metabolism and transport prediction.Trends Pharmacol Sci. 2024 Jan;45(1):39-55. doi: 10.1016/j.tips.2023.11.001. Epub 2023 Dec 9. Trends Pharmacol Sci. 2024. PMID: 38072723 Review.
-
The Use of Artificial Intelligence in Pharmacovigilance: A Systematic Review of the Literature.Pharmaceut Med. 2022 Oct;36(5):295-306. doi: 10.1007/s40290-022-00441-z. Epub 2022 Jul 29. Pharmaceut Med. 2022. PMID: 35904529
Cited by
-
Multi-task localization of the hemidiaphragms and lung segmentation in portable chest X-ray images of COVID-19 patients.Digit Health. 2024 Feb 1;10:20552076231225853. doi: 10.1177/20552076231225853. eCollection 2024 Jan-Dec. Digit Health. 2024. PMID: 38313365 Free PMC article.
-
Artificial intelligence, medications, pharmacogenomics, and ethics.Pharmacogenomics. 2024;25(14-15):611-622. doi: 10.1080/14622416.2024.2428587. Epub 2024 Nov 15. Pharmacogenomics. 2024. PMID: 39545629
-
Explainable artificial intelligence for stroke prediction through comparison of deep learning and machine learning models.Sci Rep. 2024 Dec 28;14(1):31392. doi: 10.1038/s41598-024-82931-5. Sci Rep. 2024. PMID: 39733046 Free PMC article.
-
The Impact of Artificial Intelligence on Healthcare: A Comprehensive Review of Advancements in Diagnostics, Treatment, and Operational Efficiency.Health Sci Rep. 2025 Jan 5;8(1):e70312. doi: 10.1002/hsr2.70312. eCollection 2025 Jan. Health Sci Rep. 2025. PMID: 39763580 Free PMC article.
-
Predicting Natural Product-Drug Interactions with Knowledge Graph Embeddings.AMIA Jt Summits Transl Sci Proc. 2025 Jun 10;2025:556-565. eCollection 2025. AMIA Jt Summits Transl Sci Proc. 2025. PMID: 40502231 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

