A COVID-19 DNA Vaccine Candidate Elicits Broadly Neutralizing Antibodies against Multiple SARS-CoV-2 Variants including the Currently Circulating Omicron BA.5, BF.7, BQ.1 and XBB
- PMID: 37112691
- PMCID: PMC10144402
- DOI: 10.3390/vaccines11040778
A COVID-19 DNA Vaccine Candidate Elicits Broadly Neutralizing Antibodies against Multiple SARS-CoV-2 Variants including the Currently Circulating Omicron BA.5, BF.7, BQ.1 and XBB
Abstract
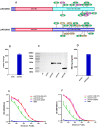
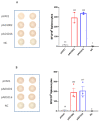
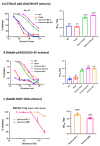
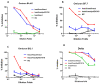
Waves of breakthrough infections by SARS-CoV-2 Omicron subvariants currently pose a global challenge to the control of the COVID-19 pandemic. We previously reported a pVAX1-based DNA vaccine candidate, pAD1002, that encodes a receptor-binding domain (RBD) chimera of SARS-CoV-1 and Omicron BA.1. In mouse and rabbit models, pAD1002 plasmid induced cross-neutralizing Abs against heterologous sarbecoviruses, including SARS-CoV-1 and SARS-CoV-2 wildtype, Delta and Omicron variants. However, these antisera failed to block the recent emerging Omicron subvariants BF.7 and BQ.1. To solve this problem, we replaced the BA.1 RBD-encoding DNA sequence in pAD1002 with that of BA.4/5. The resulting construct, namely pAD1016, elicited SARS-CoV-1 and SARS-CoV-2 RBD-specific IFN-γ+ cellular responses in BALB/c and C57BL/6 mice. More importantly, pAD1016 vaccination in mice, rabbits and pigs generated serum Abs capable of neutralizing pseudoviruses representing multiple SARS-CoV-2 Omicron subvariants including BA.2, BA.4/5, BF.7, BQ.1 and XBB. As a booster vaccine for inactivated SARS-CoV-2 virus preimmunization in mice, pAD1016 broadened the serum Ab neutralization spectrum to cover the Omicron BA.4/5, BF7 and BQ.1 subvariants. These preliminary data highlight the potential benefit of pAD1016 in eliciting neutralizing Abs against broad-spectrum Omicron subvariants in individuals previously vaccinated with inactivated prototype SARS-CoV-2 virus and suggests that pAD1016 is worthy of further translational study as a COVID-19 vaccine candidate.
Keywords: COVID-19; DNA vaccine; Omicron; RBD chimera; SARS-CoV-2.
Conflict of interest statement
All authors are Advaccine employees. G.Z., Y.D. and X.-M.G. are listed as inventors of pending patents for RBD chimera-encoding DNA vaccines.
Figures

Similar articles
-
Intranasal booster using an Omicron vaccine confers broad mucosal and systemic immunity against SARS-CoV-2 variants.Signal Transduct Target Ther. 2023 Apr 17;8(1):167. doi: 10.1038/s41392-023-01423-6. Signal Transduct Target Ther. 2023. PMID: 37069171 Free PMC article.
-
A Recombinant Protein XBB.1.5 RBD/Alum/CpG Vaccine Elicits High Neutralizing Antibody Titers against Omicron Subvariants of SARS-CoV-2.Vaccines (Basel). 2023 Oct 1;11(10):1557. doi: 10.3390/vaccines11101557. Vaccines (Basel). 2023. PMID: 37896960 Free PMC article.
-
Neutralization against Omicron subvariants after BA.5/BF.7 breakthrough infection weakened as virus evolution and aging despite repeated prototype-based vaccination1.Emerg Microbes Infect. 2023 Dec;12(2):2249121. doi: 10.1080/22221751.2023.2249121. Epub 2023 Sep 5. Emerg Microbes Infect. 2023. PMID: 37668156 Free PMC article.
-
Immune evasion of neutralizing antibodies by SARS-CoV-2 Omicron.Cytokine Growth Factor Rev. 2023 Apr;70:13-25. doi: 10.1016/j.cytogfr.2023.03.001. Epub 2023 Mar 5. Cytokine Growth Factor Rev. 2023. PMID: 36948931 Free PMC article. Review.
-
Plasma after both SARS-CoV-2 boosted vaccination and COVID-19 potently neutralizes BQ.1.1 and XBB.1.J Gen Virol. 2023 May;104(5):001854. doi: 10.1099/jgv.0.001854. J Gen Virol. 2023. PMID: 37167085 Free PMC article. Review.
Cited by
-
A review of SARS-CoV-2 variants and vaccines: Viral properties, mutations, vaccine efficacy, and safety.Infect Med (Beijing). 2023 Sep 12;2(4):247-261. doi: 10.1016/j.imj.2023.08.005. eCollection 2023 Dec. Infect Med (Beijing). 2023. PMID: 38205179 Free PMC article. Review.
-
A Three-Dose mRNA COVID-19 Vaccine Regime Produces Both Suitable Immunogenicity and Satisfactory Efficacy in Patients with Solid Cancers.Vaccines (Basel). 2023 May 23;11(6):1017. doi: 10.3390/vaccines11061017. Vaccines (Basel). 2023. PMID: 37376406 Free PMC article.
-
Retrospective study about clinical severity and epidemiological analysis of the COVID-19 Omicron subvariant lineage-infected patients in Hohhot, China.BMC Infect Dis. 2024 Feb 15;24(1):206. doi: 10.1186/s12879-024-09084-8. BMC Infect Dis. 2024. PMID: 38360539 Free PMC article.
-
Dissolving microneedles for nucleic acid delivery: A systematic search, review, and data synthesis.Acta Biomater. 2025 Jun 15;200:115-131. doi: 10.1016/j.actbio.2025.05.025. Epub 2025 May 9. Acta Biomater. 2025. PMID: 40349901 Review.
-
COVID-19 Vaccination Reporting and Adverse Event Analysis in Taiwan.Vaccines (Basel). 2024 May 29;12(6):591. doi: 10.3390/vaccines12060591. Vaccines (Basel). 2024. PMID: 38932320 Free PMC article.
References
-
- WHO Interim Statement on COVID-19 Vaccines in the Context of the Circulation of the Omicron SARS-CoV-2 Variant from the WHO Technical Advisory Group on COVID-19 Vaccine Composition (TAG-CO-VAC) 2022. [(accessed on 16 June 2022)]. Available online: https://www.who.int/news/item/11-01-2022-interim-statement-on-covid-2019....
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

