Genomic Analysis of a New Freshwater Cyanophage Lbo240-yong1 Suggests a New Taxonomic Family of Bacteriophages
- PMID: 37112811
- PMCID: PMC10140849
- DOI: 10.3390/v15040831
Genomic Analysis of a New Freshwater Cyanophage Lbo240-yong1 Suggests a New Taxonomic Family of Bacteriophages
Abstract
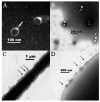
A worldwide ecological issue, cyanobacterial blooms in marine and freshwater have caused enormous losses in both the economy and the environment. Virulent cyanophages-specifically, infecting and lysing cyanobacteria-are key ecological factors involved in limiting the overall extent of the population development of cyanobacteria. Over the past three decades, reports have mainly focused on marine Prochlorococcus and Synechococcus cyanophages, while information on freshwater cyanophages remained largely unknown. In this study, a novel freshwater cyanophage, named Lbo240-yong1, was isolated via the double-layer agar plate method using Leptolyngbya boryana FACHB-240 as a host. Transmission electron microscopy observation illustrated the icosahedral head (50 ± 5 nm in diameter) and short tail (20 ± 5 nm in length) of Lbo240-yong1. Experimental infection against 37 cyanobacterial strains revealed that host-strain-specific Lbo240-yong1 could only lyse FACHB-240. The complete genome of Lbo240-yong1 is a double-stranded DNA of 39,740 bp with a G+C content of 51.99%, and it harbors 44 predicted open reading frames (ORFs). A Lbo240-yong1 ORF shared the highest identity with a gene of a filamentous cyanobacterium, hinting at a gene exchange between the cyanophage and cyanobacteria. A BLASTn search illustrated that Lbo240-yong1 had the highest sequence similarity with the Phormidium cyanophage Pf-WMP4 (89.67% identity, 84% query coverage). In the proteomic tree based on genome-wide sequence similarities, Lbo240-yong1, three Phormidium cyanophages (Pf-WMP4, Pf-WMP3, and PP), one Anabaena phage (A-4L), and one unclassified Arthronema cyanophage (Aa-TR020) formed a monophyletic group that was more deeply diverging than several other families. Pf-WMP4 is the only member of the independent genus Wumpquatrovirus that belongs to the Caudovircetes class. Pf-WMP3 and PP formed the independent genus Wumptrevirus. Anabaena phage A-4L is the only member of the independent Kozyakovvirus genus. The six cyanopodoviruses share similar gene arrangements. Eight core genes were found in them. We propose, here, to set up a new taxonomic family comprising the six freshwater cyanopodoviruses infecting filamentous cyanobacteria. This study enriched the field's knowledge of freshwater cyanophages.
Keywords: Leptolyngbya boryana; cyanophage; genome; isolation; phylogenetic tree.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Cyanophage Engineering for Algal Blooms Control.Viruses. 2024 Nov 6;16(11):1745. doi: 10.3390/v16111745. Viruses. 2024. PMID: 39599859 Free PMC article. Review.
-
A Novel Freshwater Cyanophage, Mae-Yong924-1, Reveals a New Family.Viruses. 2022 Jan 28;14(2):283. doi: 10.3390/v14020283. Viruses. 2022. PMID: 35215876 Free PMC article.
-
A Novel Freshwater Cyanophage Mae-Yong1326-1 Infecting Bloom-Forming Cyanobacterium Microcystis aeruginosa.Viruses. 2022 Sep 16;14(9):2051. doi: 10.3390/v14092051. Viruses. 2022. PMID: 36146857 Free PMC article.
-
Genomic analysis of freshwater cyanophage Pf-WMP3 Infecting cyanobacterium Phormidium foveolarum: the conserved elements for a phage.Microb Ecol. 2008 Nov;56(4):671-80. doi: 10.1007/s00248-008-9386-7. Epub 2008 Apr 29. Microb Ecol. 2008. PMID: 18443848
-
Cyanophage infection in the bloom-forming cyanobacteria Microcystis aeruginosa in surface freshwater.Microbes Environ. 2012;27(4):350-5. doi: 10.1264/jsme2.me12037. Epub 2012 Oct 5. Microbes Environ. 2012. PMID: 23047146 Free PMC article. Review.
Cited by
-
Cyanophage Engineering for Algal Blooms Control.Viruses. 2024 Nov 6;16(11):1745. doi: 10.3390/v16111745. Viruses. 2024. PMID: 39599859 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

