Avian Influenza Virus Tropism in Humans
- PMID: 37112812
- PMCID: PMC10142937
- DOI: 10.3390/v15040833
Avian Influenza Virus Tropism in Humans
Abstract
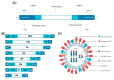
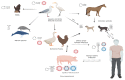
An influenza pandemic happens when a novel influenza A virus is able to infect and transmit efficiently to a new, distinct host species. Although the exact timing of pandemics is uncertain, it is known that both viral and host factors play a role in their emergence. Species-specific interactions between the virus and the host cell determine the virus tropism, including binding and entering cells, replicating the viral RNA genome within the host cell nucleus, assembling, maturing and releasing the virus to neighboring cells, tissues or organs before transmitting it between individuals. The influenza A virus has a vast and antigenically varied reservoir. In wild aquatic birds, the infection is typically asymptomatic. Avian influenza virus (AIV) can cross into new species, and occasionally it can acquire the ability to transmit from human to human. A pandemic might occur if a new influenza virus acquires enough adaptive mutations to maintain transmission between people. This review highlights the key determinants AIV must achieve to initiate a human pandemic and describes how AIV mutates to establish tropism and stable human adaptation. Understanding the tropism of AIV may be crucial in preventing virus transmission in humans and may help the design of vaccines, antivirals and therapeutic agents against the virus.
Keywords: antiviral; avian influenza virus; influenza A virus; vaccine; virus tropism in humans.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Tissue Tropisms of Avian Influenza A Viruses Affect Their Spillovers from Wild Birds to Pigs.J Virol. 2020 Nov 23;94(24):e00847-20. doi: 10.1128/JVI.00847-20. Print 2020 Nov 23. J Virol. 2020. PMID: 32967956 Free PMC article.
-
Host and viral determinants of influenza A virus species specificity.Nat Rev Microbiol. 2019 Jan;17(2):67-81. doi: 10.1038/s41579-018-0115-z. Nat Rev Microbiol. 2019. PMID: 30487536 Review.
-
Influenza A Virus Agnostic Receptor Tropism Revealed Using a Novel Biological System with Terminal Sialic Acid Knockout Cells.J Virol. 2022 Aug 10;96(15):e0041622. doi: 10.1128/jvi.00416-22. Epub 2022 Jul 18. J Virol. 2022. PMID: 35862707 Free PMC article.
-
Viral Determinants in H5N1 Influenza A Virus Enable Productive Infection of HeLa Cells.J Virol. 2020 Jan 31;94(4):e01410-19. doi: 10.1128/JVI.01410-19. Print 2020 Jan 31. J Virol. 2020. PMID: 31776276 Free PMC article.
-
[Influenza, an existing public health problem].Salud Publica Mex. 2006 May-Jun;48(3):244-67. doi: 10.1590/s0036-36342006000300009. Salud Publica Mex. 2006. PMID: 16813133 Review. Spanish.
Cited by
-
Revisiting pulmonary fibrosis: inflammatory dynamics of the lipofibroblast-to-inflammatory lipofibroblast-to-activated myofibroblast reversible switch.Front Immunol. 2025 Jun 18;16:1609509. doi: 10.3389/fimmu.2025.1609509. eCollection 2025. Front Immunol. 2025. PMID: 40607394 Free PMC article. Review.
-
Genomic Characterization and Phylogenetic Analysis of Five Avian Influenza H5N1 Subtypes from Wild Anser indicus in Yunnan, China.Vet Sci. 2025 Mar 17;12(3):280. doi: 10.3390/vetsci12030280. Vet Sci. 2025. PMID: 40267014 Free PMC article.
-
Avian Influenza Virus: Comparative Evolution as the Key for Predicting Host Tropism Expansion.Pathogens. 2025 Jun 20;14(7):608. doi: 10.3390/pathogens14070608. Pathogens. 2025. PMID: 40732656 Free PMC article.
-
An Ultra-Compact and Low-Cost LAMP-Based Virus Detection Device.Sensors (Basel). 2024 Jul 29;24(15):4912. doi: 10.3390/s24154912. Sensors (Basel). 2024. PMID: 39123959 Free PMC article.
-
Avian influenza in birds: Insights from a comprehensive review.Vet World. 2024 Nov;17(11):2544-2555. doi: 10.14202/vetworld.2024.2544-2555. Epub 2024 Nov 13. Vet World. 2024. PMID: 39829652 Free PMC article. Review.
References
-
- Kien F., Ma H.L., Bruzzone R., Poon L.L., Nal B. Definition of the cellular interactome of the highly pathogenic avian influenza H5N1 virus: Identification of human cellular regulators of viral entry, assembly, and egress. Hong Kong Med. J. 2016;22:10–12. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

