A Novel Insertion in the Hepatitis B Virus Surface Protein Leading to Hyperglycosylation Causes Diagnostic and Immune Escape
- PMID: 37112819
- PMCID: PMC10144012
- DOI: 10.3390/v15040838
A Novel Insertion in the Hepatitis B Virus Surface Protein Leading to Hyperglycosylation Causes Diagnostic and Immune Escape
Abstract
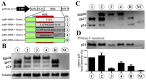
Chronic hepatitis B virus (HBV) infection is a global health threat. Mutations in the surface antigen of HBV (HBsAg) may alter its antigenicity, infectivity, and transmissibility. A patient positive for HBV DNA and detectable but low-level HBsAg in parallel with anti-HBs suggested the presence of immune and/or diagnostic escape variants. To support this hypothesis, serum-derived HBs gene sequences were amplified and cloned for sequencing, which revealed infection with exclusively non-wildtype HBV subgenotype (sgt) D3. Three distinct mutations in the antigenic loop of HBsAg that caused additional N-glycosylation were found in the variant sequences, including a previously undescribed six-nucleotide insertion. Cellular and secreted HBsAg was analyzed for N-glycosylation in Western blot after expression in human hepatoma cells. Secreted HBsAg was also subjected to four widely used, state-of-the-art diagnostic assays, which all failed to detect the hyperglycosylated insertion variant. Additionally, the recognition of mutant HBsAg by vaccine- and natural infection-induced anti-HBs antibodies was severely impaired. Taken together, these data suggest that the novel six-nucleotide insertion as well as two other previously described mutations causing hyperglycosylation in combination with immune escape mutations have a critical impact on in vitro diagnostics and likely increase the risk of breakthrough infection by evasion of vaccine-induced immunity.
Keywords: N-linked glycosylation; diagnostic escape; hepatitis B virus; immune escape.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- W.H.O . Global Progress Report on HIV, Viral Hepatitis and Sexually Transmitted Infections, 2021: Accountability for the Global Health Sector Strategies 2016–2021: Actions for Impact: Web Annex 2: Data Methods. WHO; Geneva, Switzerland: 2021.
-
- CDC . The ABCs of Hepatitis Fact Sheet. CDC; Atlanta, USA: 2013.
-
- Verrier E.R., Colpitts C.C., Bach C., Heydmann L., Weiss A., Renaud M., Durand S.C., Habersetzer F., Durantel D., Abou-Jaoude G., et al. A targeted functional RNA interference screen uncovers glypican 5 as an entry factor for hepatitis B and D viruses. Hepatology. 2016;63:35–48. doi: 10.1002/hep.28013. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

