Loss of In Vivo Replication Fitness of HIV-1 Variants Resistant to the Tat Inhibitor, dCA
- PMID: 37112931
- PMCID: PMC10146675
- DOI: 10.3390/v15040950
Loss of In Vivo Replication Fitness of HIV-1 Variants Resistant to the Tat Inhibitor, dCA
Abstract
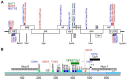
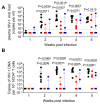
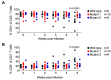
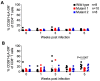
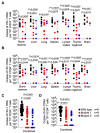
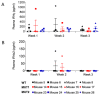
HIV resistance to the Tat inhibitor didehydro-cortistatin A (dCA) in vitro correlates with higher levels of Tat-independent viral transcription and a seeming inability to enter latency, which rendered resistant isolates more susceptible to CTL-mediated immune clearance. Here, we investigated the ability of dCA-resistant viruses to replicate in vivo using a humanized mouse model of HIV infection. Animals were infected with WT or two dCA-resistant HIV-1 isolates in the absence of dCA and followed for 5 weeks. dCA-resistant viruses exhibited lower replication rates compared to WT. Viral replication was suppressed early after infection, with viral emergence at later time points. Multiplex analysis of cytokine and chemokines from plasma samples early after infection revealed no differences in expression levels between groups, suggesting that dCA-resistance viruses did not elicit potent innate immune responses capable of blocking the establishment of infection. Viral single genome sequencing results from plasma samples collected at euthanasia revealed that at least half of the total number of mutations in the LTR region of the HIV genome considered essential for dCA evasion reverted to WT. These results suggest that dCA-resistant viruses identified in vitro suffer a fitness cost in vivo, with mutations in LTR and Nef pressured to revert to wild type.
Keywords: HIV-1; Tat inhibitor; dCA; latency promoting agent; resistance; transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Resistance to the Tat Inhibitor Didehydro-Cortistatin A Is Mediated by Heightened Basal HIV-1 Transcription.mBio. 2019 Jul 2;10(4):e01750-18. doi: 10.1128/mBio.01750-18. mBio. 2019. PMID: 31266880 Free PMC article.
-
Unexpected Mutations in HIV-1 That Confer Resistance to the Tat Inhibitor Didehydro-Cortistatin A.mBio. 2019 Jul 9;10(4):e01547-19. doi: 10.1128/mBio.01547-19. mBio. 2019. PMID: 31289189 Free PMC article.
-
The Tat Inhibitor Didehydro-Cortistatin A Prevents HIV-1 Reactivation from Latency.mBio. 2015 Jul 7;6(4):e00465. doi: 10.1128/mBio.00465-15. mBio. 2015. PMID: 26152583 Free PMC article.
-
Tat-Based Therapies as an Adjuvant for an HIV-1 Functional Cure.Viruses. 2020 Apr 8;12(4):415. doi: 10.3390/v12040415. Viruses. 2020. PMID: 32276443 Free PMC article. Review.
-
HIV Tat as a latency reversing agent: turning the tables on viral persistence.Front Immunol. 2025 Apr 11;16:1571151. doi: 10.3389/fimmu.2025.1571151. eCollection 2025. Front Immunol. 2025. PMID: 40292298 Free PMC article. Review.
Cited by
-
Analysis of the effect of HDAC inhibitors on the formation of the HIV reservoir.mBio. 2024 Sep 11;15(9):e0163224. doi: 10.1128/mbio.01632-24. Epub 2024 Aug 13. mBio. 2024. PMID: 39136440 Free PMC article.
-
Ripretinib inhibits HIV-1 transcription through modulation of PI3K-AKT-mTOR.Acta Pharmacol Sin. 2024 Aug;45(8):1632-1643. doi: 10.1038/s41401-024-01282-z. Epub 2024 Apr 16. Acta Pharmacol Sin. 2024. PMID: 38627462 Free PMC article.
-
Transient CD4+ T cell depletion during suppressive ART reduces the HIV reservoir in humanized mice.PLoS Pathog. 2023 Dec 6;19(12):e1011824. doi: 10.1371/journal.ppat.1011824. eCollection 2023 Dec. PLoS Pathog. 2023. PMID: 38055722 Free PMC article.
-
An Outbred Calf Model for Determining Innate Immune Sensing and Evolutionary Trajectories of a Cell Culture-Adapted Bovine Foamy Virus Variant.Viruses. 2023 Aug 20;15(8):1772. doi: 10.3390/v15081772. Viruses. 2023. PMID: 37632114 Free PMC article.
References
-
- Mousseau G., Clementz M.A., Bakeman W.N., Nagarsheth N., Cameron M., Shi J., Baran P., Fromentin R., Chomont N., Valente S.T. An analog of the natural steroidal alkaloid cortistatin A potently suppresses Tat-dependent HIV transcription. Cell Host Microbe. 2012;12:97–108. doi: 10.1016/j.chom.2012.05.016. - DOI - PMC - PubMed
-
- Mediouni S., Chinthalapudi K., Ekka M.K., Usui I., Jablonski J.A., Clementz M.A., Mousseau G., Nowak J., Macherla V.R., Beverage J.N., et al. Didehydro-Cortistatin A Inhibits HIV-1 by Specifically Binding to the Unstructured Basic Region of Tat. mBio. 2019;10:e02662-18. doi: 10.1128/mBio.02662-18. - DOI - PMC - PubMed
-
- Kessing C.F., Nixon C.C., Li C., Tsai P., Takata H., Mousseau G., Ho P.T., Honeycutt J.B., Fallahi M., Trautmann L., et al. In Vivo Suppression of HIV Rebound by Didehydro-Cortistatin A, a “Block-and-Lock” Strategy for HIV-1 Treatment. Cell Rep. 2017;21:600–611. doi: 10.1016/j.celrep.2017.09.080. - DOI - PMC - PubMed
-
- Mousseau G., Aneja R., Clementz M.A., Mediouni S., Lima N.S., Haregot A., Kessing C.F., Jablonski J.A., Thenin-Houssier S., Nagarsheth N., et al. Resistance to the Tat Inhibitor Didehydro-Cortistatin A Is Mediated by Heightened Basal HIV-1 Transcription. mBio. 2019;10:e01750-18. doi: 10.1128/mBio.01750-18. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

