Evaluating Novel Quantification Methods for Infectious Baculoviruses
- PMID: 37112978
- PMCID: PMC10141099
- DOI: 10.3390/v15040998
Evaluating Novel Quantification Methods for Infectious Baculoviruses
Abstract
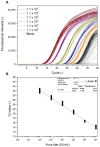
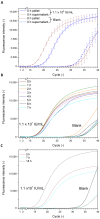
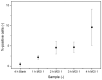
Accurate and rapid quantification of (infectious) virus titers is of paramount importance in the manufacture of viral vectors and vaccines. Reliable quantification data allow efficient process development at a laboratory scale and thorough process monitoring in later production. However, current gold standard applications, such as endpoint dilution assays, are cumbersome and do not provide true process analytical monitoring. Accordingly, flow cytometry and quantitative polymerase chain reaction have attracted increasing interest in recent years, offering various advantages for rapid quantification. Here, we compared different approaches for the assessment of infectious viruses, using a model baculovirus. Firstly, infectivity was estimated by the quantification of viral nucleic acids in infected cells, and secondly, different flow cytometric approaches were investigated regarding analysis times and calibration ranges. The flow cytometry technique included a quantification based on post-infection fluorophore expression and labeling of a viral surface protein using fluorescent antibodies. Additionally, the possibility of viral (m)RNA labeling in infected cells was investigated as a proof of concept. The results confirmed that infectivity assessment based on qPCR is not trivial and requires sophisticated method optimization, whereas staining of viral surface proteins is a fast and feasible approach for enveloped viruses. Finally, labeling of viral (m)RNA in infected cells appears to be a promising opportunity but will require further research.
Keywords: flow cytometry; gp64 protein; infectivity titer; native baculovirus; process monitoring; quantitative polymerase chain reaction; viral RNA; virus quantification.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of this study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

Similar articles
-
Quantitative real-time PCR for rapid and accurate titration of recombinant baculovirus particles.Biotechnol Bioeng. 2007 Mar 1;96(4):810-4. doi: 10.1002/bit.21177. Biotechnol Bioeng. 2007. PMID: 16952179
-
Determination of the baculovirus transducing titer in mammalian cells.Biotechnol Bioeng. 2006 Feb 20;93(3):564-71. doi: 10.1002/bit.20749. Biotechnol Bioeng. 2006. PMID: 16255040
-
Design and characterization of a compact dual channel virus counter.Cytometry A. 2005 Jun;65(2):140-7. doi: 10.1002/cyto.a.20145. Cytometry A. 2005. PMID: 15830378
-
Rapid titration of recombinant baculoviruses based on NanoLuc secretion in early infection.J Virol Methods. 2022 Sep;307:114565. doi: 10.1016/j.jviromet.2022.114565. Epub 2022 Jun 18. J Virol Methods. 2022. PMID: 35728698 Review.
-
Baculovirus vectors: novel mammalian cell gene-delivery vehicles and their applications.Am J Pharmacogenomics. 2003;3(1):53-63. Am J Pharmacogenomics. 2003. PMID: 12562216 Review.
Cited by
-
A Novel Monoclonal Antibody Against a Modified Vaccinia Ankara (MVA) Envelope Protein as a Tool for MVA Virus Titration by Flow Cytometry.Viruses. 2024 Oct 17;16(10):1628. doi: 10.3390/v16101628. Viruses. 2024. PMID: 39459960 Free PMC article.
-
The Bioengineering of Insect Cell Lines for Biotherapeutics and Vaccine Production: An Updated Review.Vaccines (Basel). 2025 May 23;13(6):556. doi: 10.3390/vaccines13060556. Vaccines (Basel). 2025. PMID: 40573887 Free PMC article. Review.
-
An automated and high-throughput approach for enhanced precision of adenoviral titering.Mol Ther Methods Clin Dev. 2025 Jan 20;33(1):101410. doi: 10.1016/j.omtm.2025.101410. eCollection 2025 Mar 13. Mol Ther Methods Clin Dev. 2025. PMID: 39968184 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

