Raman spectroscopy for viral diagnostics
- PMID: 37113565
- PMCID: PMC10088700
- DOI: 10.1007/s12551-023-01059-4
Raman spectroscopy for viral diagnostics
Abstract
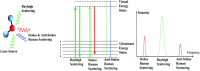
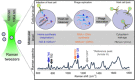
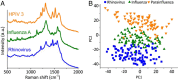
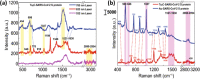
Raman spectroscopy offers the potential for fingerprinting biological molecules at ultra-low concentration and therefore has potential for the detection of viruses. Here we review various Raman techniques employed for the investigation of viruses. Different Raman techniques are discussed including conventional Raman spectroscopy, surface-enhanced Raman spectroscopy, Raman tweezer, tip-enhanced Raman Spectroscopy, and coherent anti-Stokes Raman scattering. Surface-enhanced Raman scattering can play an essential role in viral detection by multiplexing nanotechnology, microfluidics, and machine learning for ensuring spectral reproducibility and efficient workflow in sample processing and detection. The application of these techniques to diagnose the SARS-CoV-2 virus is also reviewed.
Supplementary information: The online version contains supplementary material available at 10.1007/s12551-023-01059-4.
Keywords: Point of care applications; Raman spectroscopy; Raman tweezer; SARS-CoV-2; Surface-enhanced Raman spectroscopy; Tip-enhanced Raman spectroscopy; Virus.
© The Author(s) 2023.
Conflict of interest statement
Conflict of interestThe authors declare no competing interests.
Figures









References
-
- Ali S, Hassan M, Saleem M, Tahir SF. Deep transfer learning based hepatitis B virus diagnosis using spectroscopic images. Int J Imaging Syst Technol. 2021;31:94–105. doi: 10.1002/ima.22462. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

