Comprehensive analysis of the cuproptosis-related gene DLD across cancers: A potential prognostic and immunotherapeutic target
- PMID: 37113760
- PMCID: PMC10127393
- DOI: 10.3389/fphar.2023.1111462
Comprehensive analysis of the cuproptosis-related gene DLD across cancers: A potential prognostic and immunotherapeutic target
Abstract
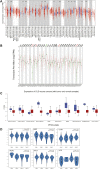
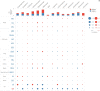
DLD is a key gene involved in "cuproptosis," but its roles in tumor progression and immunity remain unclear. Exploring the potential mechanisms and biological roles of DLD may provide new insights for therapeutic strategies for tumors. In the present study, we analyzed the role of DLD in a variety of tumors by using several bioinformatic tools. The results showed that compared with normal tissues, tumor tissues representing multiple cancers showed significant differential expression of DLD. High DLD expression was associated with a good prognosis in BRCA, KICH, and LUAD. Conversely, high expression levels of DLD were detrimental to patient prognosis in many other tumors, such as COAD, KIRC, and KIRP. In addition, the associations of DLD with infiltrating immune cells, genetic alterations and methylation levels across cancers were assessed. Aberrant expression of DLD was positively correlated with most infiltrating immune cells, especially neutrophils. The DLD methylation level was significantly decreased in COAD, LIHC, and LUSC but significantly increased in BRCA. DLD had the highest mutation rate (6.04%) in ESCA. In LUSC, patients with genetic alterations in DLD showed a poorer prognosis. At the single-cell level, the roles of DLD in regulating cancer-associated biological functions, such as metastasis, inflammation, and differentiation, were explored. Afterward, we further investigated whether several disease-associated genes could be correlated with DLD. GO enrichment analysis indicated that DLD-related genes were mainly associated with mitochondria-related cellular components, aerobic respiration and the tricarboxylic acid cycle. Finally, the correlations between DLD expression and immunomodulatory genes, immune checkpoints, and sensitivity to some antitumor drugs were investigated. It is worth noting that DLD expression was positively correlated with immune checkpoint genes and immunomodulatory genes in most cancers. In conclusion, this study comprehensively analyzed the differential expression, prognostic value and immune cell infiltration-related function of DLD across cancers. Our results suggest that DLD has great potential to serve as a candidate marker for pancancer prognosis and immunotherapy and may provide a new direction for cancer treatment development.
Keywords: bioinformatics analysis; cuproptosis; dld; immune cell infiltration; pan-cancer; prognosis.
Copyright © 2023 Yang, Guo, Wu, Tong, Xiao, Wang, Liu, Xu, Yan and Sun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures













References
-
- Ambrus A., Wang J., Mizsei R., Zambo Z., Torocsik B., Jordan F., et al. (2016). Structural alterations induced by ten disease-causing mutations of human dihydrolipoamide dehydrogenase analyzed by hydrogen/deuterium-exchange mass spectrometry: Implications for the structural basis of E3 deficiency. Biochim. Biophys. Acta 1862 (11), 2098–2109. 10.1016/j.bbadis.2016.08.013 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

