Parallel phospholipid transfer by Vps13 and Atg2 determines autophagosome biogenesis dynamics
- PMID: 37115156
- PMCID: PMC10148235
- DOI: 10.1083/jcb.202211039
Parallel phospholipid transfer by Vps13 and Atg2 determines autophagosome biogenesis dynamics
Abstract
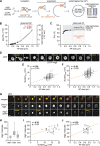
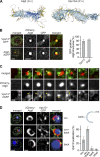
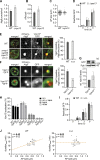
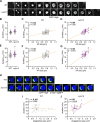
During autophagy, rapid membrane assembly expands small phagophores into large double-membrane autophagosomes. Theoretical modeling predicts that the majority of autophagosomal phospholipids are derived from highly efficient non-vesicular phospholipid transfer (PLT) across phagophore-ER contacts (PERCS). Currently, the phagophore-ER tether Atg2 is the only PLT protein known to drive phagophore expansion in vivo. Here, our quantitative live-cell imaging analysis reveals a poor correlation between the duration and size of forming autophagosomes and the number of Atg2 molecules at PERCS of starving yeast cells. Strikingly, we find that Atg2-mediated PLT is non-rate limiting for autophagosome biogenesis because membrane tether and the PLT protein Vps13 localizes to the rim and promotes the expansion of phagophores in parallel with Atg2. In the absence of Vps13, the number of Atg2 molecules at PERCS determines the duration and size of forming autophagosomes with an apparent in vivo transfer rate of ∼200 phospholipids per Atg2 molecule and second. We propose that conserved PLT proteins cooperate in channeling phospholipids across organelle contact sites for non-rate-limiting membrane assembly during autophagosome biogenesis.
© 2023 Dabrowski et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures



References
-
- Anding, A.L., Wang C., Chang T.K., Sliter D.A., Powers C.M., Hofmann K., Youle R.J., and Baehrecke E.H.. 2018. Vps13D encodes a ubiquitin-binding protein that is required for the regulation of mitochondrial size and clearance. Curr. Biol. 28:287–295.e6. 10.1016/j.cub.2017.11.064 - DOI - PMC - PubMed
-
- Axe, E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., and Ktistakis N.T.. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701. 10.1083/jcb.200803137 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

