An excavate root for the eukaryote tree of life
- PMID: 37115919
- PMCID: PMC10146883
- DOI: 10.1126/sciadv.ade4973
An excavate root for the eukaryote tree of life
Abstract
Much of the higher-order phylogeny of eukaryotes is well resolved, but the root remains elusive. We assembled a dataset of 183 eukaryotic proteins of archaeal ancestry to test this root. The resulting phylogeny identifies four lineages of eukaryotes currently classified as "Excavata" branching separately at the base of the tree. Thus, Parabasalia appear as the first major branch of eukaryotes followed sequentially by Fornicata, Preaxostyla, and Discoba. All four excavate branch points receive full statistical support from analyses with commonly used evolutionary models, a protein structure partition model that we introduce here, and various controls for deep phylogeny artifacts. The absence of aerobic mitochondria in Parabasalia, Fornicata, and Preaxostyla suggests that modern eukaryotes arose under anoxic conditions, probably much earlier than expected, and without the benefit of mitochondrial respiration.
Figures

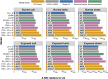
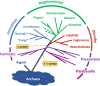
References
-
- Burki F., Roger A. J., Brown M. W., Simpson A. G. B., The new tree of eukaryotes. Trends Ecol. Evol. 35, 43–55 (2020). - PubMed
-
- Simpson A. G. B., Cytoskeletal organization, phylogenetic affinities and systematics in the contentious taxon Excavata (Eukaryota). Int. J. Syst. Evol. Microbiol. 53, 1759–1777 (2003). - PubMed
-
- Adl S. M., Bass D., Lane C. E., Lukeš J., Schoch C. L., Smirnov A., Agatha S., Berney C., Brown M. W., Burki F., Cárdenas P., Čepička I., Chistyakova L., del Campo J., Dunthorn M., Edvardsen B., Eglit Y., Guillou L., Hampl V., Heiss A. A., Hoppenrath M., James T. Y., Karnkowska A., Karpov S., Kim E., Kolisko M., Kudryavtsev A., Lahr D. J. G., Lara E., Le Gall L., Lynn D. H., Mann D. G., Massana R., Mitchell E. A. D., Morrow C., Park J. S., Pawlowski J. W., Powell M. J., Richter D. J., Rueckert S., Shadwick L., Shimano S., Spiegel F. W., Torruella G., Youssef N., Zlatogursky V., Zhang Q., Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 66, 4–119 (2019). - PMC - PubMed
-
- Burger G., Valach M., Perfection of eccentricity: Mitochondrial genomes of diplonemids. IUBMB Life 70, 1197–1206 (2018). - PubMed
-
- Van Der Giezen M., Hydrogenosomes and Mitosomes: Conservation and evolution of functions. J. Eukaryot. Microbiol. 56, 221–231 (2009). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

