Potential use of high-resolution melting analyses for SARS-CoV-2 genomic surveillance
- PMID: 37116586
- PMCID: PMC10132831
- DOI: 10.1016/j.jviromet.2023.114742
Potential use of high-resolution melting analyses for SARS-CoV-2 genomic surveillance
Abstract
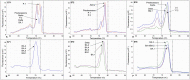
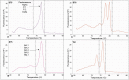
The pandemic caused by COVID-19 and the emergence of new variants of SARS-CoV-2 have generated clinical and epidemiological impacts on a global scale. The use of strategies for monitoring viral circulation and identifying mutations in genomic regions involved in host interaction are important measures to mitigate viral dissemination and reduce its likely complications on population health. In this context, the objective of this work was to explore the potential of high-resolution melting (HRM) analysis combined with one-step real-time reverse transcription PCR in a closed-tube system, as a fast and convenient method of screening for SARS-CoV-2 mutations with possible implications on host-pathogen interactions. The HRM analyses allowed the distinction of the Gamma, Zeta, Alpha, Delta, and Omicron variants against the predecessors (B.1.1.28, B.1.1.33) of occurrence in Brazil. It is concluded that the molecular tool standardized here has the potential to optimize the genomic surveillance of SARS-CoV-2, and could be adapted for genomic surveillance of other pathogens, due to its ability to detect, prior to sequencing, samples suggestive of new variants, selecting them more assertively and earlier for whole genome sequencing when compared to random screening.
Keywords: COVID-19; Genomic sequencing; HRM; SARS-CoV-2; Variants.
Copyright © 2023. Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
References
-
- ARTIC Network, 2020. artic-network/primer-schemes: v1.1.1. https://doi.org/10.5281/ZENODO.4020380.
-
- ARTIC Network, 2021. SARS-CoV-2 V4.1 update for Omicron variant - Laboratory - ARTIC Real-time Genomic Surveillance [WWW Document]. URL 〈https://community.artic.network/t/sars-cov-2-v4–1-update-for-omicron-va... (accessed 1.15.23).
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous