Vector Aided Microenvironment programming (VAMP): reprogramming the TME with MVA virus expressing IL-12 for effective antitumor activity
- PMID: 37117006
- PMCID: PMC10151998
- DOI: 10.1136/jitc-2023-006718
Vector Aided Microenvironment programming (VAMP): reprogramming the TME with MVA virus expressing IL-12 for effective antitumor activity
Abstract
Background: Tumor microenvironment (TME) represents a critical hurdle in cancer immunotherapy, given its ability to suppress antitumor immunity. Several efforts are made to overcome this hostile TME with the development of new therapeutic strategies modifying TME to boost antitumor immunity. Among these, cytokine-based approaches have been pursued for their known immunomodulatory effects on different cell populations within the TME. IL-12 is a potent pro-inflammatory cytokine that demonstrates striking immune activation and tumor control but causes severe adverse effects when systemically administered. Thus, local administration is considered a potential strategy to achieve high cytokine concentrations at the tumor site while sparing systemic adverse effects.
Methods: Modified Vaccinia Ankara (MVA) vector is a potent inducer of pro-inflammatory response. Here, we cloned IL-12 into the genome of MVA for intratumoral immunotherapy, combining the immunomodulatory properties of both the vector and the cargo. The antitumor activity of MVA-IL-12 and its effect on TME reprogramming were investigated in preclinical tumor models. RNA sequencing (RNA-Seq) analysis was performed to assess changes in the TME in treated and distal tumors and the effect on the intratumoral T-cell receptor repertoire.
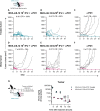
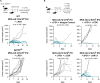
Results: Intratumoral injection of MVA-IL-12 resulted in strong antitumor activity with the complete remission of established tumors in multiple murine models, including those resistant to checkpoint inhibitors. The therapeutic activity of MVA-IL-12 was associated with very low levels of circulating cytokine. Effective TME reprogramming was demonstrated on treatment, with the reduction of immunosuppressive M2 macrophages while increasing pro-inflammatory M1, and recruitment of dendritic cells. TME switch from immunosuppressive into immunostimulatory environment allowed for CD8 T cells priming and expansion leading to tumor attack.
Conclusions: Intratumoral administration of MVA-IL-12 turns immunologically 'cold' tumors 'hot' and overcomes resistance to programmed cell death protein-1 blockade.
Keywords: cytokines; immunomodulation; immunotherapy; lymphocytes, tumor-infiltrating; tumor microenvironment.
© Author(s) (or their employer(s)) 2023. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: ES is founder of Nouscom. AMD, LS, LI, LN, MDL, GC, GL, EM, IG, GS, FT, SA, GR are employees of Nouscom.
Figures





Similar articles
-
Intratumoral expression of interleukin 23 variants using oncolytic vaccinia virus elicit potent antitumor effects on multiple tumor models via tumor microenvironment modulation.Theranostics. 2021 May 3;11(14):6668-6681. doi: 10.7150/thno.56494. eCollection 2021. Theranostics. 2021. PMID: 34093846 Free PMC article.
-
TG6050, an oncolytic vaccinia virus encoding interleukin-12 and anti-CTLA-4 antibody, favors tumor regression via profound immune remodeling of the tumor microenvironment.J Immunother Cancer. 2024 Jul 25;12(7):e009302. doi: 10.1136/jitc-2024-009302. J Immunother Cancer. 2024. PMID: 39060022 Free PMC article.
-
Intratumoral virotherapy with 4-1BBL armed modified vaccinia Ankara eradicates solid tumors and promotes protective immune memory.J Immunother Cancer. 2021 Feb;9(2):e001586. doi: 10.1136/jitc-2020-001586. J Immunother Cancer. 2021. PMID: 33579736 Free PMC article.
-
Reprogramming the breast tumor immune microenvironment: cold-to-hot transition for enhanced immunotherapy.J Exp Clin Cancer Res. 2025 Apr 25;44(1):131. doi: 10.1186/s13046-025-03394-8. J Exp Clin Cancer Res. 2025. PMID: 40281554 Free PMC article. Review.
-
CD8+ cytotoxic T lymphocytes in cancer immunotherapy: A review.J Cell Physiol. 2019 Jun;234(6):8509-8521. doi: 10.1002/jcp.27782. Epub 2018 Nov 22. J Cell Physiol. 2019. PMID: 30520029 Review.
Cited by
-
Interleukin-12 Delivery Strategies and Advances in Tumor Immunotherapy.Curr Issues Mol Biol. 2024 Oct 16;46(10):11548-11579. doi: 10.3390/cimb46100686. Curr Issues Mol Biol. 2024. PMID: 39451566 Free PMC article. Review.
References
-
- Bejarano L, Jordāo MJC, Joyce JA. Therapeutic targeting of the tumor microenvironment. Cancer Discov 2021;11:933–59. 10.1158/2159-8290.CD-20-1808 - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
