Dependency of NELF-E-SLUG-KAT2B epigenetic axis in breast cancer carcinogenesis
- PMID: 37117180
- PMCID: PMC10147683
- DOI: 10.1038/s41467-023-38132-1
Dependency of NELF-E-SLUG-KAT2B epigenetic axis in breast cancer carcinogenesis
Abstract
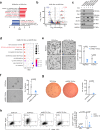
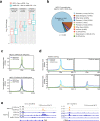
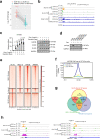
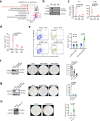
Cancer cells undergo transcriptional reprogramming to drive tumor progression and metastasis. Using cancer cell lines and patient-derived tumor organoids, we demonstrate that loss of the negative elongation factor (NELF) complex inhibits breast cancer development through downregulating epithelial-mesenchymal transition (EMT) and stemness-associated genes. Quantitative multiplexed Rapid Immunoprecipitation Mass spectrometry of Endogenous proteins (qPLEX-RIME) further reveals a significant rewiring of NELF-E-associated chromatin partners as a function of EMT and a co-option of NELF-E with the key EMT transcription factor SLUG. Accordingly, loss of NELF-E leads to impaired SLUG binding on chromatin. Through integrative transcriptomic and genomic analyses, we identify the histone acetyltransferase, KAT2B, as a key functional target of NELF-E-SLUG. Genetic and pharmacological inactivation of KAT2B ameliorate the expression of EMT markers, phenocopying NELF ablation. Elevated expression of NELF-E and KAT2B is associated with poorer prognosis in breast cancer patients, highlighting the clinical relevance of our findings. Taken together, we uncover a crucial role of the NELF-E-SLUG-KAT2B epigenetic axis in breast cancer carcinogenesis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

