MasitinibL shows promise as a drug-like analog of masitinib that elicits comparable SARS-Cov-2 3CLpro inhibition with low kinase preference
- PMID: 37117213
- PMCID: PMC10141821
- DOI: 10.1038/s41598-023-33024-2
MasitinibL shows promise as a drug-like analog of masitinib that elicits comparable SARS-Cov-2 3CLpro inhibition with low kinase preference
Abstract
SARS-CoV-2 infection has led to several million deaths worldwide and ravaged the economies of many countries. Hence, developing therapeutics against SARS-CoV-2 remains a core priority in the fight against COVID-19. Most of the drugs that have received emergency use authorization for treating SARS-CoV-2 infection exhibit a number of limitations, including side effects and questionable efficacy. This challenge is further compounded by reinfection after vaccination and the high likelihood of mutations, as well as the emergence of viral escape mutants that render SARS-CoV-2 spike glycoprotein-targeting vaccines ineffective. Employing de novo drug synthesis or repurposing to discover broad-spectrum antivirals that target highly conserved pathways within the viral machinery is a focus of current research. In a recent drug repurposing study, masitinib, a clinically safe drug against the human coronavirus OC43 (HCoV-OC43), was identified as an antiviral agent with effective inhibitory activity against the SARS-CoV-2 3CLpro. Masitinib is currently under clinical trial in combination with isoquercetin in hospitalized patients (NCT04622865). Nevertheless, masitinib has kinase-related side effects; hence, the development of masitinib analogs with lower anti-tyrosine kinase activity becomes necessary. In this study, in an attempt to address this limitation, we executed a comprehensive virtual workflow in silico to discover drug-like compounds matching selected pharmacophore features in the SARS-CoV-2 3CLpro-bound state of masitinib. We identified a novel lead compound, "masitinibL", a drug-like analog of masitinib that demonstrated strong inhibitory properties against the SARS-CoV-2 3CLpro. In addition, masitinibL further displayed low selectivity for tyrosine kinases, which strongly suggests that masitinibL is a highly promising therapeutic that is preferable to masitinib.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




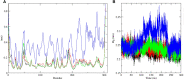




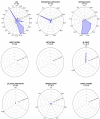
References
-
- Wong CK, et al. Real-world effectiveness of early molnupiravir or nirmatrelvir–ritonavir in hospitalised patients with COVID-19 without supplemental oxygen requirement on admission during Hong Kong’s omicron BA. 2 wave: A retrospective cohort study. Lancet Infect. Dis. 2022;22(12):1681–1693. doi: 10.1016/S1473-3099(22)00507-2. - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

