The burden of rare protein-truncating genetic variants on human lifespan
- PMID: 37117740
- PMCID: PMC10154195
- DOI: 10.1038/s43587-022-00182-3
The burden of rare protein-truncating genetic variants on human lifespan
Abstract
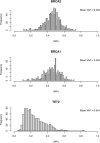
Genetic predisposition has been shown to contribute substantially to the age at which we die. Genome-wide association studies (GWASs) have linked more than 20 loci to phenotypes related to human lifespan1. However, little is known about how lifespan is impacted by gene loss of function. Through whole-exome sequencing of 352,338 UK Biobank participants of European ancestry, we assessed the relevance of protein-truncating variant (PTV) gene burden on individual and parental survival. We identified four exome-wide significant (P < 4.2 × 10-7) human lifespan genes, BRCA1, BRCA2, ATM and TET2. Gene and gene-set, PTV-burden, phenome-wide association studies support known roles of these genes in cancer to impact lifespan at the population level. The TET2 PTV burden was associated with a lifespan through somatic mutation events presumably due to clonal hematopoiesis. The overlap between PTV burden and common variant-based lifespan GWASs was modest, underscoring the value of exome sequencing in well-powered biobank cohorts to complement GWASs for identifying genes underlying complex traits.
© 2022. The Author(s).
Conflict of interest statement
J.Z.L., C.Y.C., E.A.T., C.D.W., D.S., S.J. and H.R. are full-time employees and hold stocks and stock options at Biogen.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

