SALT OVERLY SENSITIVE2 stabilizes phytochrome-interacting factors PIF4 and PIF5 to promote Arabidopsis shade avoidance
- PMID: 37119311
- PMCID: PMC10396385
- DOI: 10.1093/plcell/koad119
SALT OVERLY SENSITIVE2 stabilizes phytochrome-interacting factors PIF4 and PIF5 to promote Arabidopsis shade avoidance
Abstract
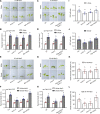
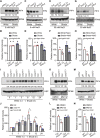
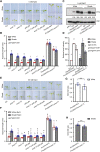
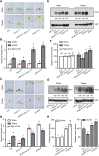
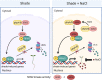
Sun-loving plants trigger the shade avoidance syndrome (SAS) to compete against their neighbors for sunlight. Phytochromes are plant red (R) and far-red (FR) light photoreceptors that play a major role in perceiving the shading signals and triggering SAS. Shade induces a reduction in the level of active phytochrome B (phyB), thus increasing the abundance of PHYTOCHROME-INTERACTING FACTORS (PIFs), a group of growth-promoting transcription factors. However, whether other factors are involved in modulating PIF activity in the shade remains largely obscure. Here, we show that SALT OVERLY SENSITIVE2 (SOS2), a protein kinase essential for salt tolerance, positively regulates SAS in Arabidopsis thaliana. SOS2 directly phosphorylates PIF4 and PIF5 at a serine residue close to their conserved motif for binding to active phyB. This phosphorylation thus decreases their interaction with phyB and posttranslationally promotes PIF4 and PIF5 protein accumulation. Notably, the role of SOS2 in regulating PIF4 and PIF5 protein abundance and SAS is more prominent under salt stress. Moreover, phyA and phyB physically interact with SOS2 and promote SOS2 kinase activity in the light. Collectively, our study uncovers an unexpected role of salt-activated SOS2 in promoting SAS by modulating the phyB-PIF module, providing insight into the coordinated response of plants to salt stress and shade.
© American Society of Plant Biologists 2023. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. The authors declare no conflict of interests.
Figures



References
-
- Bauer D, Viczián A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adám E, Fejes E, Schäfer E, et al. . Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 2004:16(6):1433–1445. 10.1105/tpc.021568 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

