Oncohistones and disrupted development in pediatric-type diffuse high-grade glioma
- PMID: 37119408
- PMCID: PMC10441521
- DOI: 10.1007/s10555-023-10105-2
Oncohistones and disrupted development in pediatric-type diffuse high-grade glioma
Abstract
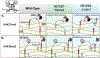
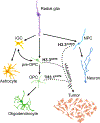
Recurrent, clonal somatic mutations in histone H3 are molecular hallmarks that distinguish the genetic mechanisms underlying pediatric and adult high-grade glioma (HGG), define biological subgroups of diffuse glioma, and highlight connections between cancer, development, and epigenetics. These oncogenic mutations in histones, now termed "oncohistones", were discovered through genome-wide sequencing of pediatric diffuse high-grade glioma. Up to 80% of diffuse midline glioma (DMG), including diffuse intrinsic pontine glioma (DIPG) and diffuse glioma arising in other midline structures including thalamus or spinal cord, contain histone H3 lysine 27 to methionine (K27M) mutations or, rarely, other alterations that result in a depletion of H3K27me3 similar to that induced by H3 K27M. This subgroup of glioma is now defined as diffuse midline glioma, H3K27-altered. In contrast, histone H3 Gly34Arg/Val (G34R/V) mutations are found in approximately 30% of diffuse glioma arising in the cerebral hemispheres of older adolescents and young adults, now classified as diffuse hemispheric glioma, H3G34-mutant. Here, we review how oncohistones modulate the epigenome and discuss the mutational landscape and invasive properties of histone mutant HGGs of childhood. The distinct mechanisms through which oncohistones and other mutations rewrite the epigenetic landscape provide novel insights into development and tumorigenesis and may present unique vulnerabilities for pHGGs. Lessons learned from these rare incurable brain tumors of childhood may have broader implications for cancer, as additional high- and low-frequency oncohistone mutations have been identified in other tumor types.
Keywords: Diffuse hemispheric glioma; Diffuse midline glioma; Epigenetic; H3 K27M; H3G34-mutant; H3K27-altered; Oncohistones; Pediatric diffuse glioma.
© 2023. The Author(s), under exclusive licence to Springer Science+Business Media, LLC, part of Springer Nature.
Conflict of interest statement
Statements and Declarations
Authors have no conflicts of interest to disclose.
Figures
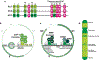

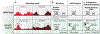


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

