Rare disease research resources at the Rat Genome Database
- PMID: 37119810
- PMCID: PMC10411567
- DOI: 10.1093/genetics/iyad078
Rare disease research resources at the Rat Genome Database
Abstract
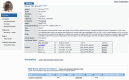
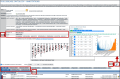
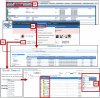
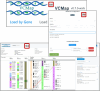
Rare diseases individually affect relatively few people, but as a group they impact considerable numbers of people. The Rat Genome Database (https://rgd.mcw.edu) is a knowledgebase that offers resources for rare disease research. This includes disease definitions, genes, quantitative trail loci (QTLs), genetic variants, annotations to published literature, links to external resources, and more. One important resource is identifying relevant cell lines and rat strains that serve as models for disease research. Diseases, genes, and strains have report pages with consolidated data, and links to analysis tools. Utilizing these globally accessible resources for rare disease research, potentiating discovery of mechanisms and new treatments, can point researchers toward solutions to alleviate the suffering of those afflicted with these diseases.
Keywords: Rat Genome Database; ontologies; rare diseases.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflicts of interest statement The author(s) declare no conflict of interest.
Figures




References
-
- Fang EF, Kassahun H, Croteau DL, Scheibye-Knudsen M, Marosi K, Lu H, Shamanna RA, Kalyanasundaram S, Bollineni RC, Wilson MA, et al. . Nad(+) replenishment improves lifespan and healthspan in ataxia telangiectasia models via mitophagy and DNA repair. Cell Metab. 2016;24(4):566–581. doi:10.1016/j.cmet.2016.09.004. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

