PARPs and ADP-ribosylation: Deciphering the complexity with molecular tools
- PMID: 37119811
- PMCID: PMC10202152
- DOI: 10.1016/j.molcel.2023.04.009
PARPs and ADP-ribosylation: Deciphering the complexity with molecular tools
Abstract
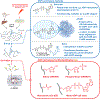
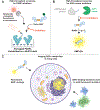
PARPs catalyze ADP-ribosylation-a post-translational modification that plays crucial roles in biological processes, including DNA repair, transcription, immune regulation, and condensate formation. ADP-ribosylation can be added to a wide range of amino acids with varying lengths and chemical structures, making it a complex and diverse modification. Despite this complexity, significant progress has been made in developing chemical biology methods to analyze ADP-ribosylated molecules and their binding proteins on a proteome-wide scale. Additionally, high-throughput assays have been developed to measure the activity of enzymes that add or remove ADP-ribosylation, leading to the development of inhibitors and new avenues for therapy. Real-time monitoring of ADP-ribosylation dynamics can be achieved using genetically encoded reporters, and next-generation detection reagents have improved the precision of immunoassays for specific forms of ADP-ribosylation. Further development and refinement of these tools will continue to advance our understanding of the functions and mechanisms of ADP-ribosylation in health and disease.
Keywords: ADP-ribose biosensor; ADP-ribosylation; ADP-ribosylome; PAR-binding proteins; PARPs; chemical biology; drug development; proteomics.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.K.L.L. holds a patent related to the ELTA technology used for labeling ADP-ribosylated molecules.
Figures


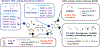


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

