A high quality, high molecular weight DNA extraction method for PacBio HiFi genome sequencing of recalcitrant plants
- PMID: 37120601
- PMCID: PMC10148486
- DOI: 10.1186/s13007-023-01009-x
A high quality, high molecular weight DNA extraction method for PacBio HiFi genome sequencing of recalcitrant plants
Abstract
Background: PacBio HiFi sequencing provides highly accurate long-read sequencing datasets which are of great advantage for whole genome sequencing projects. One limitation of the method is the requirement for high quality, high molecular weight input DNA. This can be particularly challenging for plants that frequently contain common and species-specific secondary metabolites, which often interfere with downstream processes. Cape Primroses (genus Streptocarpus), are some of these recalcitrant plants and are selected here as material to develop a high quality, high molecular weight DNA extraction protocol for long read genome sequencing.
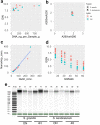
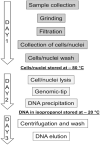
Results: We developed a DNA extraction method for PacBio HiFi sequencing for Streptocarpus grandis and Streptocarpus kentaniensis. A CTAB lysis buffer was employed to avoid guanidine, and the traditional chloroform and phenol purification steps were replaced with pre-lysis sample washes. Best cells/nucleus lysis was achieved with 4 h at 58 °C. The obtained high quality and high molecular weight DNAs were tested in PacBio SMRTBell™ library preparations, which resulted in circular consensus sequencing (CCS) reads from 17 to 27 Gb per cell, and a read length N50 from 14 to 17 kbp. To evaluate the quality of the reads for whole genome sequencing, they were assembled with HiFiasm into draft genomes, with N50 = 49 Mb and 23 Mb, and L50 = 10 and 11. The longest contigs were 95 Mb and 57 Mb respectively, showing good contiguity as these are longer than the theoretical chromosome length (genome size/chromosome number) of 78 Mb and 55 Mb, for S. grandis and S. kentaniensis respectively.
Conclusions: DNA extraction is a critical step towards obtaining a complete genome assembly. Our DNA extraction method here provided the required high quality, high molecular weight DNA for successful standard-input PacBio HiFi library preparation. The contigs from those reads showed a high contiguity, providing a good starting draft assembly towards obtaining a complete genome. The results obtained here were highly promising, and demonstrated that the DNA extraction method developed here is compatible with PacBio HiFi sequencing and suitable for de novo whole genome sequencing projects of plants.
Keywords: DNA extraction; Genome sequencing; Gesneriaceae; Long-read sequencing; Next-generation sequencing; PacBio HiFi; SMRTbell™ library; Streptocarpus.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- PacBio. Technical overview: HiFi library preparation using SMRTbell express template prep kit 2.0 for de novo assembly and variant detection applications. 2021. HiFi Library Preparation Using SMRTbell Express TPK 2.0 for De Novo Assembly and Variant Detection. https://www.pacb.com/wp-content/uploads/HiFi-Library-Preparation-Using-S....
-
- PacBio. Technical note Preparing DNA for PacBio HiFi sequencing—extraction and quality control. 2022. https://www.pacb.com/wp-content/uploads/Technical-Note-Preparing-DNA-for....
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

