Carbon-Centered Radicals in Protein Manipulation
- PMID: 37122447
- PMCID: PMC10141601
- DOI: 10.1021/acscentsci.3c00051
Carbon-Centered Radicals in Protein Manipulation
Abstract
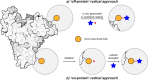
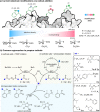
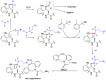
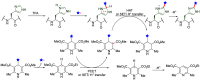
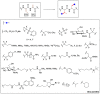
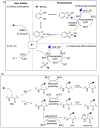
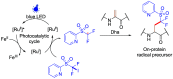
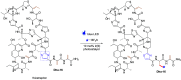
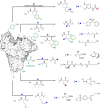
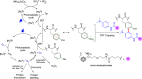
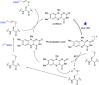
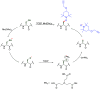
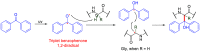
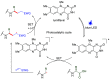
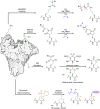
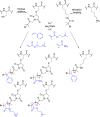
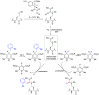
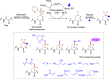
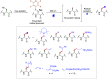
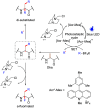
Methods to directly post-translationally modify proteins are perhaps the most straightforward and operationally simple ways to create and study protein post-translational modifications (PTMs). However, precisely altering or constructing the C-C scaffolds pervasive throughout biology is difficult with common two-electron chemical approaches. Recently, there has been a surge of new methods that have utilized single electron/radical chemistry applied to site-specifically "edit" proteins that have started to create this potential-one that in principle could be near free-ranging. This review provides an overview of current methods that install such "edits", including those that generate function and/or PTMs, through radical C-C bond formation (as well as C-X bond formation via C• where illustrative). These exploit selectivity for either native residues, or preinstalled noncanonical protein side-chains with superior radical generating or accepting abilities. Particular focus will be on the radical generation approach (on-protein or off-protein, use of light and photocatalysts), judging the compatibility of conditions with proteins and cells, and novel chemical biology applications afforded by these methods. While there are still many technical hurdles, radical C-C bond formation on proteins is a promising and rapidly growing area in chemical biology with long-term potential for biological editing.
© 2023 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Hoyt E. A.; Cal P. M.; Oliveira B. L.; Bernardes G. J. Contemporary approaches to site-selective protein modification. Nature Reviews Chemistry 2019, 3 (3), 147–171. 10.1038/s41570-019-0079-1. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

