Gene length is a pivotal feature to explain disparities in transcript capture between single transcriptome techniques
- PMID: 37122996
- PMCID: PMC10132733
- DOI: 10.3389/fbinf.2023.1144266
Gene length is a pivotal feature to explain disparities in transcript capture between single transcriptome techniques
Abstract
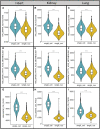
The scale and capability of single-cell and single-nucleus RNA-sequencing technologies are rapidly growing, enabling key discoveries and large-scale cell mapping operations. However, studies directly comparing technical differences between single-cell and single-nucleus RNA sequencing are still lacking. Here, we compared three paired single-cell and single-nucleus transcriptomes from three different organs (Heart, Lung and Kidney). Differently from previous studies that focused on cell classification, we explored disparities in the transcriptome output of whole cells relative to the nucleus. We found that the major cell clusters could be recovered by either technique from matched samples, but at different proportions. In 2/3 datasets (kidney and lung) we detected clusters exclusively present with single-nucleus RNA sequencing. In all three organ groups, we found that genomic and gene structural characteristics such as gene length and exon content significantly differed between the two techniques. Genes recovered with the single-nucleus RNA sequencing technique had longer sequence lengths and larger exon counts, whereas single-cell RNA sequencing captured short genes at higher rates. Furthermore, we found that when compared to the whole host genome (mouse for kidney and lung datasets and human for the heart dataset), single transcriptomes obtained with either technique skewed from the expected proportions in several points: a) coding sequence length, b) transcript length and c) genomic span; and d) distribution of genes based on exons counts. Interestingly, the top-100 DEG between the two techniques returned distinctive GO terms. Hence, the type of single transcriptome technique used affected the outcome of downstream analysis. In summary, our data revealed both techniques present disparities in RNA capture. Moreover, the biased RNA capture affected the calculations of basic cellular parameters, raising pivotal points about the limitations and advantages of either single transcriptome techniques.
Keywords: biased gene capture; bioinformatic; data analysis; next-generation sequencing; single-cell RNA sequencing; single-nucleus RNA sequencing.
Copyright © 2023 Pavan, Diniz, El-Dahr and Tortelote.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
LinkOut - more resources
Full Text Sources

