Identification and characterization of the CDK1-BMAL1-UHRF1 pathway driving tumor progression
- PMID: 37123229
- PMCID: PMC10130925
- DOI: 10.1016/j.isci.2023.106544
Identification and characterization of the CDK1-BMAL1-UHRF1 pathway driving tumor progression
Abstract
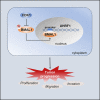
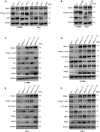
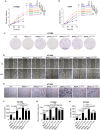
The abnormal regulation of BMAL1 could lead to the occurrence and progression of various tumors. However, the mechanism of phosphorylation regulation of BMAL1 in tumorigenesis remains poorly understood. In this study, we report a previously unrecognized BMAL1 dephosphorylation pathway that promotes tumor progression. BMAL1 accelerates cell proliferation, migration, and invasion of HT1080 and Calu1 cells. CDK1 binds to BMAL1 through a conserved domain and regulates the dephosphorylation of BMAL1 on Ser42 residues, but not on Ser78 or Thr224, thereby enhancing the oncogenic activity of BMAL1. Dephosphorylation of BMAL1 Ser42 promotes tumor growth and metastasis in mouse subcutaneous transplantation tumor and lung metastatic tumor models. Moreover, UHRF1 is recognized as an important target gene of BMAL1 in cancer cells. Consequently, UHRF1 depletion mimics BMAL1 deficiency with respect to tumor suppression, whereas transfection-enforced re-expression of UHRF1 restores tumor growth in BMAL1-deficient cells. These findings suggest a link between the circadian clock regulator and cancer progression.
Keywords: Cancer; Cell biology; Molecular biology.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Early J.O., Menon D., Wyse C.A., Cervantes-Silva M.P., Zaslona Z., Carroll R.G., Palsson-McDermott E.M., Angiari S., Ryan D.G., Corcoran S.E., et al. Circadian clock protein BMAL1 regulates IL-1β in macrophages via NRF2. Proc. Natl. Acad. Sci. USA. 2018;115:E8460–E8468. doi: 10.1073/pnas.1800431115. - DOI - PMC - PubMed
-
- Fuhr L., El-Athman R., Scrima R., Cela O., Carbone A., Knoop H., Li Y., Hoffmann K., Laukkanen M.O., Corcione F., et al. The circadian clock regulates metabolic phenotype rewiring via HKDC1 and modulates tumor progression and drug response in colorectal cancer. EBioMedicine. 2018;33:105–121. doi: 10.1016/j.ebiom.2018.07.002. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

