Rare variant enrichment analysis supports GREB1L as a contributory driver gene in the etiology of Mayer-Rokitansky-Küster-Hauser syndrome
- PMID: 37124138
- PMCID: PMC10130500
- DOI: 10.1016/j.xhgg.2023.100188
Rare variant enrichment analysis supports GREB1L as a contributory driver gene in the etiology of Mayer-Rokitansky-Küster-Hauser syndrome
Abstract
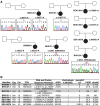
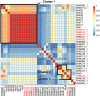
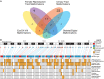
Mayer-Rokitansky-Küster-Hauser (MRKH) syndrome is characterized by aplasia of the female reproductive tract; the syndrome can include renal anomalies, absence or dysgenesis, and skeletal anomalies. While functional models have elucidated several candidate genes, only WNT4 (MIM: 603490) variants have been definitively associated with a subtype of MRKH with hyperandrogenism (MIM: 158330). DNA from 148 clinically diagnosed MRKH probands across 144 unrelated families and available family members from North America, Europe, and South America were exome sequenced (ES) and by family-based genomics analyzed for rare likely deleterious variants. A replication cohort consisting of 442 Han Chinese individuals with MRKH was used to further reproduce GREB1L findings in diverse genetic backgrounds. Proband and OMIM phenotypes annotated using the Human Phenotype Ontology were analyzed to quantitatively delineate the phenotypic spectrum associated with GREB1L variant alleles found in our MRKH cohort and those previously published. This study reports 18 novel GREB1L variant alleles, 16 within a multiethnic MRKH cohort and two within a congenital scoliosis cohort. Cohort-wide analyses for a burden of rare variants within a single gene identified likely damaging variants in GREB1L (MIM: 617782), a known disease gene for renal hypoplasia and uterine abnormalities (MIM: 617805), in 16 of 590 MRKH probands. GREB1L variant alleles, including a CNV null allele, were found in 8 MRKH type 1 probands and 8 MRKH type II probands. This study used quantitative phenotypic analyses in a worldwide multiethnic cohort to identify and strengthen the association of GREB1L to isolated uterine agenesis (MRKH type I) and syndromic MRKH type II.
Keywords: Alu-Alu mediated rearrangement; developmental genomics; exonic deletion CNV; genitourinary development; human phenotype ontology; infertility; rare variant gene enrichment; sex-limited trait; transcriptional regulation.
© 2023 The Author(s).
Conflict of interest statement
J.R.L. has stock ownership in 23andMe and is a co-inventor on multiple U.S. and European patents related to molecular diagnostics for inherited neuropathies, genomic disorders, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis and clinical genomic sequencing (both ES and WGS) offered in the Baylor Genetics Laboratory (http://bmgl.com). J.R.L. and I.B.V. serve on the Scientific Advisory Board of BG. S.E.A. is the co-founder and CEO of Medigenome, The Swiss Institute of Genomic Medicine.
Figures



References
-
- Herlin M., Bjørn A.M.B., Rasmussen M., Trolle B., Petersen M.B. Prevalence and patient characteristics of Mayer-Rokitansky-Küster-Hauser syndrome: a nationwide registry-based study. Hum. Reprod. 2016;31:2384–2390. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

