RecF protein targeting to postreplication (daughter strand) gaps I: DNA binding by RecF and RecFR
- PMID: 37125642
- PMCID: PMC10287957
- DOI: 10.1093/nar/gkad311
RecF protein targeting to postreplication (daughter strand) gaps I: DNA binding by RecF and RecFR
Erratum in
-
Correction to 'RecF protein targeting to postreplication (daughter strand) gaps I: DNA binding by RecF and RecFR'.Nucleic Acids Res. 2023 Jun 23;51(11):5900. doi: 10.1093/nar/gkad475. Nucleic Acids Res. 2023. PMID: 37216600 Free PMC article. No abstract available.
Abstract
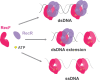
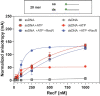
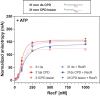
In bacteria, the repair of post-replication gaps by homologous recombination requires the action of the recombination mediator proteins RecF, RecO and RecR. Whereas the role of the RecOR proteins to displace the single strand binding protein (SSB) and facilitate RecA loading is clear, how RecF mediates targeting of the system to appropriate sites remains enigmatic. The most prominent hypothesis relies on specific RecF binding to gap ends. To test this idea, we present a detailed examination of RecF and RecFR binding to more than 40 DNA substrates of varying length and structure. Neither RecF nor the RecFR complex exhibited specific DNA binding that can explain the targeting of RecF(R) to post-replication gaps. RecF(R) bound to dsDNA and ssDNA of sufficient length with similar facility. DNA binding was highly ATP-dependent. Most measured Kd values fell into a range of 60-180 nM. The addition of ssDNA extensions on duplex substrates to mimic gap ends or CPD lesions produces only subtle increases or decreases in RecF(R) affinity. Significant RecFR binding cooperativity was evident with many DNA substrates. The results indicate that RecF or RecFR targeting to post-replication gaps must rely on factors not yet identified, perhaps involving interactions with additional proteins.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

