Megalin, cubilin, and Dab2 drive endocytic flux in kidney proximal tubule cells
- PMID: 37126375
- PMCID: PMC10295476
- DOI: 10.1091/mbc.E22-11-0510
Megalin, cubilin, and Dab2 drive endocytic flux in kidney proximal tubule cells
Abstract
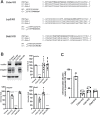
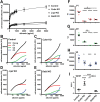
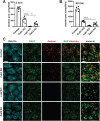
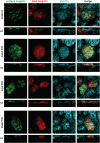
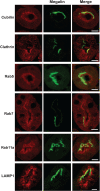
The kidney proximal tubule (PT) elaborates a uniquely high-capacity apical endocytic pathway to retrieve albumin and other proteins that escape the glomerular filtration barrier. Megalin and cubilin/amnionless (CUBAM) receptors engage Dab2 in these cells to mediate clathrin-dependent uptake of filtered ligands. Knockout of megalin or Dab2 profoundly inhibits apical endocytosis and is believed to atrophy the endocytic pathway. We generated CRISPR/Cas9 knockout (KO) clones lacking cubilin, megalin, or Dab2 expression in highly differentiated PT cells and determined the impact on albumin internalization and endocytic pathway function. KO of each component had different effects on the concentration dependence of albumin uptake as well its distribution within PT cells. Reduced uptake of a fluid phase marker was also observed, with megalin KO cells having the most dramatic decline. Surprisingly, protein levels and distribution of key endocytic proteins were preserved in KO PT cell lines and in megalin KO mice, despite the reduced endocytic activity. Our data highlight specific functions of megalin, cubilin, and Dab2 in apical endocytosis and demonstrate that these proteins drive endocytic flux without compromising the physical integrity of the apical endocytic pathway. Our studies suggest a novel model to explain how these components coordinate endocytic uptake in PT cells.
Figures



References
-
- Christensen EI, Birn H, Storm T, Weyer K, Nielsen R (2012). Endocytic receptors in the renal proximal tubule. Physiology (Bethesda) 27, 223–236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

