The genetic determinants of recurrent somatic mutations in 43,693 blood genomes
- PMID: 37126548
- PMCID: PMC10132750
- DOI: 10.1126/sciadv.abm4945
The genetic determinants of recurrent somatic mutations in 43,693 blood genomes
Abstract
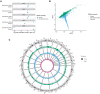
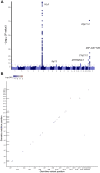
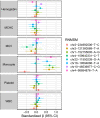
Nononcogenic somatic mutations are thought to be uncommon and inconsequential. To test this, we analyzed 43,693 National Heart, Lung and Blood Institute Trans-Omics for Precision Medicine blood whole genomes from 37 cohorts and identified 7131 non-missense somatic mutations that are recurrently mutated in at least 50 individuals. These recurrent non-missense somatic mutations (RNMSMs) are not clearly explained by other clonal phenomena such as clonal hematopoiesis. RNMSM prevalence increased with age, with an average 50-year-old having 27 RNMSMs. Inherited germline variation associated with RNMSM acquisition. These variants were found in genes involved in adaptive immune function, proinflammatory cytokine production, and lymphoid lineage commitment. In addition, the presence of eight specific RNMSMs associated with blood cell traits at effect sizes comparable to Mendelian genetic mutations. Overall, we found that somatic mutations in blood are an unexpectedly common phenomenon with ancestry-specific determinants and human health consequences.
Figures

References
-
- K. Yizhak, F. Aguet, J. Kim, J. M. Hess, K. Kübler, J. Grimsby, R. Frazer, H. Zhang, N. J. Haradhvala, D. Rosebrock, D. Livitz, X. Li, E. Arich-Landkof, N. Shoresh, C. Stewart, A. V. Segrè, P. A. Branton, P. Polak, K. G. Ardlie, G. Getz, RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 364, eaaw0726 (2019). - PMC - PubMed
-
- S. Abelson, G. Collord, S. W. K. Ng, O. Weissbrod, N. Mendelson Cohen, E. Niemeyer, N. Barda, P. C. Zuzarte, L. Heisler, Y. Sundaravadanam, R. Luben, S. Hayat, T. T. Wang, Z. Zhao, I. Cirlan, T. J. Pugh, D. Soave, K. Ng, C. Latimer, C. Hardy, K. Raine, D. Jones, D. Hoult, A. Britten, J. D. McPherson, M. Johansson, F. Mbabaali, J. Eagles, J. K. Miller, D. Pasternack, L. Timms, P. Krzyzanowski, P. Awadalla, R. Costa, E. Segal, S. V. Bratman, P. Beer, S. Behjati, I. Martincorena, J. C. Y. Wang, K. M. Bowles, J. R. Quirós, A. Karakatsani, C. La Vecchia, A. Trichopoulou, E. Salamanca-Fernández, J. M. Huerta, A. Barricarte, R. C. Travis, R. Tumino, G. Masala, H. Boeing, S. Panico, R. Kaaks, A. Krämer, S. Sieri, E. Riboli, P. Vineis, M. Foll, J. McKay, S. Polidoro, N. Sala, K.-T. Khaw, R. Vermeulen, P. J. Campbell, E. Papaemmanuil, M. D. Minden, A. Tanay, R. D. Balicer, N. J. Wareham, M. Gerstung, J. E. Dick, P. Brennan, G. S. Vassiliou, L. I. Shlush, Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559, 400–404 (2018). - PMC - PubMed
-
- S. Jaiswal, P. Fontanillas, J. Flannick, A. Manning, P. V. Grauman, B. G. Mar, R. Coleman Lindsley, C. H. Mermel, N. Burtt, A. Chavez, J. M. Higgins, V. Moltchanov, F. C. Kuo, M. J. Kluk, B. Henderson, L. Kinnunen, H. A. Koistinen, C. Ladenvall, G. Getz, A. Correa, B. F. Banahan, S. Gabriel, S. Kathiresan, H. M. Stringham, M. I. McCarthy, M. Boehnke, J. Tuomilehto, C. Haiman, L. Groop, G. Atzmon, J. G. Wilson, D. Neuberg, D. Altshuler, B. L. Ebert, Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 26, 2488–2998 (2014). - PMC - PubMed
-
- G. Genovese, A. K. Kähler, R. E. Handsaker, J. Lindberg, S. A. Rose, S. F. Bakhoum, K. Chambert, E. Mick, B. M. Neale, M. Fromer, S. M. Purcell, O. Svantesson, M. Landén, M. Höglund, S. Lehmann, S. B. Gabriel, J. L. Moran, E. S. Lander, P. F. Sullivan, P. Sklar, H. Grönberg, C. M. Hultman, S. A. McCarroll, Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014). - PMC - PubMed

