Small open reading frames: a comparative genetics approach to validation
- PMID: 37127568
- PMCID: PMC10152738
- DOI: 10.1186/s12864-023-09311-7
Small open reading frames: a comparative genetics approach to validation
Abstract
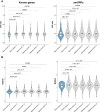
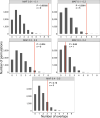
Open reading frames (ORFs) with fewer than 100 codons are generally not annotated in genomes, although bona fide genes of that size are known. Newer biochemical studies have suggested that thousands of small protein-coding ORFs (smORFs) may exist in the human genome, but the true number and the biological significance of the micropeptides they encode remain uncertain. Here, we used a comparative genomics approach to identify high-confidence smORFs that are likely protein-coding. We identified 3,326 high-confidence smORFs using constraint within human populations and evolutionary conservation as additional lines of evidence. Next, we validated that, as a group, our high-confidence smORFs are conserved at the amino-acid level rather than merely residing in highly conserved non-coding regions. Finally, we found that high-confidence smORFs are enriched among disease-associated variants from GWAS. Overall, our results highlight that smORF-encoded peptides likely have important functional roles in human disease.
Keywords: Comparative genetics; Evolutionary conservation; Human genetic variation; Micropeptides; Small open reading frames.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Stein CS, Jadiya P, Zhang X, McLendon JM, Abouassaly GM, Witmer NH, Anderson EJ, Elrod JW, Boudreau RL. Mitoregulin: A lncRNA-Encoded Microprotein that Supports Mitochondrial Supercomplexes and Respiratory Efficiency. Cell Rep. 2018;23(13):3710–3720e3718. doi: 10.1016/j.celrep.2018.06.002. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

