Dynamic epistasis analysis reveals how chromatin remodeling regulates transcriptional bursting
- PMID: 37127821
- PMCID: PMC10191856
- DOI: 10.1038/s41594-023-00981-1
Dynamic epistasis analysis reveals how chromatin remodeling regulates transcriptional bursting
Abstract
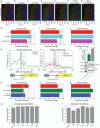
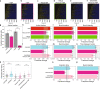
Transcriptional bursting has been linked to the stochastic positioning of nucleosomes. However, how bursting is regulated by the remodeling of promoter nucleosomes is unknown. Here, we use single-molecule live-cell imaging of GAL10 transcription in Saccharomyces cerevisiae to measure how bursting changes upon combined perturbations of chromatin remodelers, the transcription factor Gal4 and preinitiation complex components. Using dynamic epistasis analysis, we reveal how the remodeling of different nucleosomes regulates transcriptional bursting parameters. At the nucleosome covering the Gal4 binding sites, RSC and Gal4 binding synergistically facilitate each burst. Conversely, nucleosome remodeling at the TATA box controls only the first burst upon galactose induction. At canonical TATA boxes, the nucleosomes are displaced by TBP binding to allow for transcription activation even in the absence of remodelers. Overall, our results reveal how promoter nucleosome remodeling together with Gal4 and preinitiation complex binding regulates transcriptional bursting.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures













References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

