Applying causal discovery to single-cell analyses using CausalCell
- PMID: 37129360
- PMCID: PMC10229139
- DOI: 10.7554/eLife.81464
Applying causal discovery to single-cell analyses using CausalCell
Abstract
Correlation between objects is prone to occur coincidentally, and exploring correlation or association in most situations does not answer scientific questions rich in causality. Causal discovery (also called causal inference) infers causal interactions between objects from observational data. Reported causal discovery methods and single-cell datasets make applying causal discovery to single cells a promising direction. However, evaluating and choosing causal discovery methods and developing and performing proper workflow remain challenges. We report the workflow and platform CausalCell (http://www.gaemons.net/causalcell/causalDiscovery/) for performing single-cell causal discovery. The workflow/platform is developed upon benchmarking four kinds of causal discovery methods and is examined by analyzing multiple single-cell RNA-sequencing (scRNA-seq) datasets. Our results suggest that different situations need different methods and the constraint-based PC algorithm with kernel-based conditional independence tests work best in most situations. Related issues are discussed and tips for best practices are given. Inferred causal interactions in single cells provide valuable clues for investigating molecular interactions and gene regulations, identifying critical diagnostic and therapeutic targets, and designing experimental and clinical interventions.
Keywords: causal analysis; causal relationship; computational biology; feature selection; human; mouse; network inference; scRNA-seq; single-cell analysis; systems biology.
© 2023, Wen, Huang et al.
Conflict of interest statement
YW, JH, SG, YE, AM, HZ, YD, HZ No competing interests declared
Figures


















































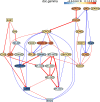








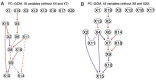
Update of
References
-
- Alban TJ, Bayik D, Otvos B, Rabljenovic A, Leng L, Jia-Shiun L, Roversi G, Lauko A, Momin AA, Mohammadi AM, Peereboom DM, Ahluwalia MS, Matsuda K, Yun K, Bucala R, Vogelbaum MA, Lathia JD. Glioblastoma myeloid-derived suppressor cell subsets express differential macrophage migration inhibitory factor receptor profiles that can be targeted to reduce immune suppression. Frontiers in Immunology. 2020;11:1191. doi: 10.3389/fimmu.2020.01191. - DOI - PMC - PubMed
-
- Angelidis I, Simon LM, Fernandez IE, Strunz M, Mayr CH, Greiffo FR, Tsitsiridis G, Ansari M, Graf E, Strom T-M, Nagendran M, Desai T, Eickelberg O, Mann M, Theis FJ, Schiller HB. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nature Communications. 2019;10:963. doi: 10.1038/s41467-019-08831-9. - DOI - PMC - PubMed
-
- Avelar RA, Ortega JG, Tacutu R, Tyler EJ, Bennett D, Binetti P, Budovsky A, Chatsirisupachai K, Johnson E, Murray A, Shields S, Tejada-Martinez D, Thornton D, Fraifeld VE, Bishop CL, de Magalhães JP. A multidimensional systems biology analysis of cellular senescence in aging and disease. Genome Biology. 2020;21:91. doi: 10.1186/s13059-020-01990-9. - DOI - PMC - PubMed
-
- Balcazar N, Sathyamurthy A, Elghazi L, Gould A, Weiss A, Shiojima I, Walsh K, Bernal-Mizrachi E. Mtorc1 activation regulates beta-cell mass and proliferation by modulation of cyclin D2 synthesis and stability. The Journal of Biological Chemistry. 2009;284:7832–7842. doi: 10.1074/jbc.M807458200. - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

