Machine learning-driven multifunctional peptide engineering for sustained ocular drug delivery
- PMID: 37130851
- PMCID: PMC10154330
- DOI: 10.1038/s41467-023-38056-w
Machine learning-driven multifunctional peptide engineering for sustained ocular drug delivery
Abstract
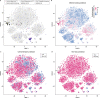
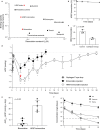
Sustained drug delivery strategies have many potential benefits for treating a range of diseases, particularly chronic diseases that require treatment for years. For many chronic ocular diseases, patient adherence to eye drop dosing regimens and the need for frequent intraocular injections are significant barriers to effective disease management. Here, we utilize peptide engineering to impart melanin binding properties to peptide-drug conjugates to act as a sustained-release depot in the eye. We develop a super learning-based methodology to engineer multifunctional peptides that efficiently enter cells, bind to melanin, and have low cytotoxicity. When the lead multifunctional peptide (HR97) is conjugated to brimonidine, an intraocular pressure lowering drug that is prescribed for three times per day topical dosing, intraocular pressure reduction is observed for up to 18 days after a single intracameral injection in rabbits. Further, the cumulative intraocular pressure lowering effect increases ~17-fold compared to free brimonidine injection. Engineered multifunctional peptide-drug conjugates are a promising approach for providing sustained therapeutic delivery in the eye and beyond.
© 2023. The Author(s).
Conflict of interest statement
H.T.H, R.T.C., J.H., M.P.C., and L.M.E. are named as inventors on the U.S. Provisional Patent Application No. 63/340,714, which covers aspects of this work, and has been jointly filed by the Johns Hopkins University and University of Maryland, College Park. The other authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

