Mutations in α-synuclein, TDP-43 and tau prolong protein half-life through diminished degradation by lysosomal proteases
- PMID: 37131250
- PMCID: PMC10155372
- DOI: 10.1186/s13024-023-00621-8
Mutations in α-synuclein, TDP-43 and tau prolong protein half-life through diminished degradation by lysosomal proteases
Erratum in
-
Correction: Mutations in α-synuclein, TDP-43 and tau prolong protein half-life through diminished degradation by lysosomal proteases.Mol Neurodegener. 2023 May 30;18(1):34. doi: 10.1186/s13024-023-00629-0. Mol Neurodegener. 2023. PMID: 37254128 Free PMC article. No abstract available.
Abstract
Background: Autosomal dominant mutations in α-synuclein, TDP-43 and tau are thought to predispose to neurodegeneration by enhancing protein aggregation. While a subset of α-synuclein, TDP-43 and tau mutations has been shown to increase the structural propensity of these proteins toward self-association, rates of aggregation are also highly dependent on protein steady state concentrations, which are in large part regulated by their rates of lysosomal degradation. Previous studies have shown that lysosomal proteases operate precisely and not indiscriminately, cleaving their substrates at very specific linear amino acid sequences. With this knowledge, we hypothesized that certain coding mutations in α-synuclein, TDP-43 and tau may lead to increased protein steady state concentrations and eventual aggregation by an alternative mechanism, that is, through disrupting lysosomal protease cleavage recognition motifs and subsequently conferring protease resistance to these proteins.
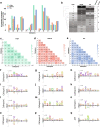
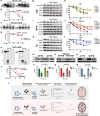
Results: To test this possibility, we first generated comprehensive proteolysis maps containing all of the potential lysosomal protease cleavage sites for α-synuclein, TDP-43 and tau. In silico analyses of these maps indicated that certain mutations would diminish cathepsin cleavage, a prediction we confirmed utilizing in vitro protease assays. We then validated these findings in cell models and induced neurons, demonstrating that mutant forms of α-synuclein, TDP-43 and tau are degraded less efficiently than wild type despite being imported into lysosomes at similar rates.
Conclusions: Together, this study provides evidence that pathogenic mutations in the N-terminal domain of α-synuclein (G51D, A53T), low complexity domain of TDP-43 (A315T, Q331K, M337V) and R1 and R2 domains of tau (K257T, N279K, S305N) directly impair their own lysosomal degradation, altering protein homeostasis and increasing cellular protein concentrations by extending the degradation half-lives of these proteins. These results also point to novel, shared, alternative mechanism by which different forms of neurodegeneration, including synucleinopathies, TDP-43 proteinopathies and tauopathies, may arise. Importantly, they also provide a roadmap for how the upregulation of particular lysosomal proteases could be targeted as potential therapeutics for human neurodegenerative disease.
Keywords: Autophagy; Cathepsin; Lysosome; Mutations; Neurodegeneration; Protease; TDP-43; Tau; α-synuclein.
© 2023. The Author(s).
Conflict of interest statement
A.W.K. and M.P.J. are Scientific Advisory Board members, and M.P.J. is a founder of Nine Square Therapeutics. C.S.C. is a founder, and G.M.K. is an employee of Alaunus Biosciences. Authors declare that they have no conflicts of interest or competing interests.
Figures