Crohn's disease proteolytic microbiota enhances inflammation through PAR2 pathway in gnotobiotic mice
- PMID: 37131291
- PMCID: PMC10158566
- DOI: 10.1080/19490976.2023.2205425
Crohn's disease proteolytic microbiota enhances inflammation through PAR2 pathway in gnotobiotic mice
Abstract
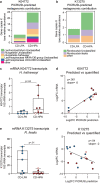
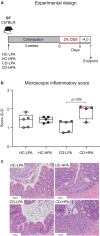
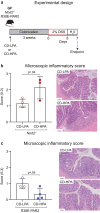
Emerging evidence implicates microbial proteolytic activity in ulcerative colitis (UC), but whether it also plays a role in Crohn's disease (CD) remains unclear. We investigated the effects of colonizing adult and neonatal germ-free C57BL/6 mice with CD microbiota, selected based on high (CD-HPA) or low fecal proteolytic activity (CD-LPA), or microbiota from healthy controls with LPA (HC-LPA) or HPA (HC-HPA). We then investigated colitogenic mechanisms in gnotobiotic C57BL/6, and in mice with impaired Nucleotide-binding Oligomerization Domain-2 (NOD2) and Protease-Activated Receptor 2 (PAR2) cleavage resistant mice (Nod2-/-; R38E-PAR2 respectively). At sacrifice, total fecal proteolytic, elastolytic, and mucolytic activity were analyzed. Microbial community and predicted function were assessed by 16S rRNA gene sequencing and PICRUSt2. Immune function and colonic injury were investigated by inflammatory gene expression (NanoString) and histology. Colonization with HC-LPA or CD-LPA lowered baseline fecal proteolytic activity in germ-free mice, which was paralleled by lower acute inflammatory cell infiltrate. CD-HPA further increased proteolytic activity compared with germ-free mice. CD-HPA mice had lower alpha diversity, distinct microbial profiles and higher fecal proteolytic activity compared with CD-LPA. C57BL/6 and Nod2-/- mice, but not R38E-PAR2, colonized with CD-HPA had higher colitis severity than those colonized with CD-LPA. Our results indicate that CD proteolytic microbiota is proinflammatory, increasing colitis severity through a PAR2 pathway.
Keywords: Crohn’s disease; DSS-induced colitis; Proteinase-activated receptor 2 (PAR2); gnotobiotic mice; inflammation; proteolytic activity.
Conflict of interest statement
EFV is Member of the Biocodex International and National (Canada) Scientific Review Boards, Member of the Center for Gut Microbiome Research and Education Scientific Advisory Board of the AGA, Secretary of the International Society of the Study of Celiac Disease, and holds grants from Kallyope and Codexis, unrelated to this study.
Figures



References
-
- Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390(10114):2769–2778. doi:10.1016/S0140-6736(17)32448-0. - DOI - PubMed
-
- Motta J, Magne L, Descamps D, Rolland C, Squarzoni–Dale C, Rousset P, Martin L, Cenac N, Balloy V, Huerre M, et al. Modifying the protease, antiprotease pattern by Elafin overexpression protects mice from colitis. Gastroenterology. 2011;140(4):1272–1282. doi:10.1053/j.gastro.2010.12.050. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
