Identification of Ferroptosis-Related Biomarkers for Diagnosis and Molecular Classification of Staphylococcus aureus-Induced Osteomyelitis
- PMID: 37131411
- PMCID: PMC10149083
- DOI: 10.2147/JIR.S406562
Identification of Ferroptosis-Related Biomarkers for Diagnosis and Molecular Classification of Staphylococcus aureus-Induced Osteomyelitis
Abstract
Objective: Staphylococcus aureus (SA)-induced osteomyelitis (OM) is one of the most common refractory diseases in orthopedics. Early diagnosis is beneficial to improve the prognosis of patients. Ferroptosis plays a key role in inflammation and immune response, while the mechanism of ferroptosis-related genes (FRGs) in SA-induced OM is still unclear. The purpose of this study was to determine the role of ferroptosis-related genes in the diagnosis, molecular classification and immune infiltration of SA-induced OM by bioinformatics.
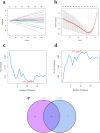
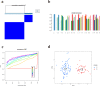
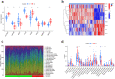
Methods: Datasets related to SA-induced OM and ferroptosis were collected from the Gene Expression Omnibus (GEO) and ferroptosis databases, respectively. The least absolute shrinkage and selection operator (LASSO) and support vector machine-recursive feature elimination (SVM-RFE) algorithms were combined to screen out differentially expressed-FRGs (DE-FRGs) with diagnostic characteristics, and gene set enrichment analysis (GSEA) and gene set variation analysis (GSVA) were used to explore specific biological functions and pathways. Based on these key DE-FRGs, a diagnostic model was established, and molecular subtypes were divided to explore the changes in the immune microenvironment between molecular subtypes.
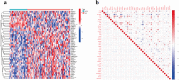
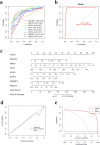
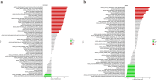
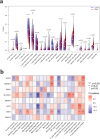
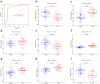
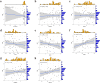
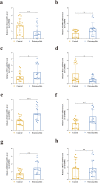
Results: A total of 41 DE-FRGs were identified. After screening and intersecting with LASSO and SVM-RFE algorithms, 8 key DE-FRGs with diagnostic characteristics were obtained, which may regulate the pathogenesis of OM through the immune response and amino acid metabolism. The ROC curve indicated that the 8 DE-FRGs had excellent diagnostic ability for SA-induced OM (AUC=0.993). Two different molecular subtypes (subtype 1 and subtype 2) were identified by unsupervised cluster analysis. The CIBERSORT analysis revealed that the subtype 1 OM had higher immune cell infiltration rates, mainly in T cells CD4 memory resting, macrophages M0, macrophages M2, dendritic cells resting, and dendritic cells activated.
Conclusion: We developed a diagnostic model related to ferroptosis and molecular subtypes significantly related to immune infiltration, which may provide a novel insight for exploring the pathogenesis and immunotherapy of SA-induced OM.
Keywords: biomarker; ferroptosis; immune infiltration; molecular subtype; osteomyelitis.
© 2023 Shi et al.
Conflict of interest statement
All the authors declare that they have no competing interests in this work.
Figures


References
-
- Momodu II, Savaliya V. Osteomyelitis. Treasure Island (FL): StatPearls Publishing Copyright © 2022, StatPearls Publishing LLC; 2022.
LinkOut - more resources
Full Text Sources
Research Materials

