This is a preprint.
The hagfish genome and the evolution of vertebrates
- PMID: 37131617
- PMCID: PMC10153176
- DOI: 10.1101/2023.04.17.537254
The hagfish genome and the evolution of vertebrates
Update in
-
The hagfish genome and the evolution of vertebrates.Nature. 2024 Mar;627(8005):811-820. doi: 10.1038/s41586-024-07070-3. Epub 2024 Jan 23. Nature. 2024. PMID: 38262590 Free PMC article.
Abstract
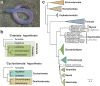
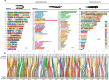
As the only surviving lineages of jawless fishes, hagfishes and lampreys provide a critical window into early vertebrate evolution. Here, we investigate the complex history, timing, and functional role of genome-wide duplications in vertebrates in the light of a chromosome-scale genome of the brown hagfish Eptatretus atami. Using robust chromosome-scale (paralogon-based) phylogenetic methods, we confirm the monophyly of cyclostomes, document an auto-tetraploidization (1RV) that predated the origin of crown group vertebrates ~517 Mya, and establish the timing of subsequent independent duplications in the gnathostome and cyclostome lineages. Some 1RV gene duplications can be linked to key vertebrate innovations, suggesting that this early genomewide event contributed to the emergence of pan-vertebrate features such as neural crest. The hagfish karyotype is derived by numerous fusions relative to the ancestral cyclostome arrangement preserved by lampreys. These genomic changes were accompanied by the loss of genes essential for organ systems (eyes, osteoclast) that are absent in hagfish, accounting in part for the simplification of the hagfish body plan; other gene family expansions account for hagfishes' capacity to produce slime. Finally, we characterise programmed DNA elimination in somatic cells of hagfish, identifying protein-coding and repetitive elements that are deleted during development. As in lampreys, the elimination of these genes provides a mechanism for resolving genetic conflict between soma and germline by repressing germline/pluripotency functions. Reconstruction of the early genomic history of vertebrates provides a framework for further exploration of vertebrate novelties.
Figures



References
-
- Altenhoff AM, Levy J, Zarowiecki M, Tomiczek B, Warwick Vesztrocy A, Dalquen DA, Müller S, Telford MJ, Glover NM, Dylus D, Dessimoz C. 2019. OMA standalone: orthology inference among public and custom genomes and transcriptomes. Genome Res 29:1152–1163. doi:10.1101/gr.243212.118 - DOI - PMC - PubMed
-
- Chen Z, Omori Y, Koren S, Shirokiya T, Kuroda T, Miyamoto A, Wada H, Fujiyama A, Toyoda A, Zhang S, Wolfsberg TG, Kawakami K, Phillippy AM, NISC Comparative Sequencing Program, Mullikin JC, Burgess SM. 2019. De novo assembly of the goldfish (Carassius auratus) genome and the evolution of genes after whole-genome duplication. Sci Adv 5:eaav0547. doi:10.1126/sciadv.aav0547 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
