This is a preprint.
Gene activity fully predicts transcriptional bursting dynamics
- PMID: 37131882
- PMCID: PMC10153294
Gene activity fully predicts transcriptional bursting dynamics
Abstract
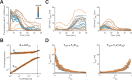
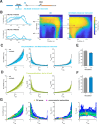
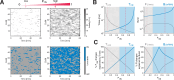
Transcription commonly occurs in bursts, with alternating productive (ON) and quiescent (OFF) periods, governing mRNA production rates. Yet, how transcription is regulated through bursting dynamics remains unresolved. Here, we conduct real-time measurements of endogenous transcriptional bursting with single-mRNA sensitivity. Leveraging the diverse transcriptional activities in early fly embryos, we uncover stringent relationships between bursting parameters. Specifically, we find that the durations of ON and OFF periods are linked. Regardless of the developmental stage or body-axis position, gene activity levels predict individual alleles' average ON and OFF periods. Lowly transcribing alleles predominantly modulate OFF periods (burst frequency), while highly transcribing alleles primarily tune ON periods (burst size). These relationships persist even under perturbations of cis-regulatory elements or trans-factors and account for bursting dynamics measured in other species. Our results suggest a novel mechanistic constraint governing bursting dynamics rather than a modular control of distinct parameters by distinct regulatory processes.
Figures



Similar articles
-
A conserved coupling of transcriptional ON and OFF periods underlies bursting dynamics.Nat Struct Mol Biol. 2025 Jul 15. doi: 10.1038/s41594-025-01615-4. Online ahead of print. Nat Struct Mol Biol. 2025. PMID: 40664961
-
Unified bursting strategies in ectopic and endogenous even-skipped expression patterns.bioRxiv [Preprint]. 2024 Jun 25:2023.02.09.527927. doi: 10.1101/2023.02.09.527927. bioRxiv. 2024. Update in: Elife. 2024 Dec 09;12:RP88671. doi: 10.7554/eLife.88671. PMID: 36798351 Free PMC article. Updated. Preprint.
-
Transcriptional bursting dynamics in gene expression.Front Genet. 2024 Sep 13;15:1451461. doi: 10.3389/fgene.2024.1451461. eCollection 2024. Front Genet. 2024. PMID: 39346775 Free PMC article. Review.
-
What shapes eukaryotic transcriptional bursting?Mol Biosyst. 2017 Jun 27;13(7):1280-1290. doi: 10.1039/c7mb00154a. Mol Biosyst. 2017. PMID: 28573295 Review.
-
Differential context-specific impact of individual core promoter elements on transcriptional dynamics.Mol Biol Cell. 2017 Nov 7;28(23):3360-3370. doi: 10.1091/mbc.E17-06-0408. Epub 2017 Sep 20. Mol Biol Cell. 2017. PMID: 28931597 Free PMC article.
References
-
- Cramer P., Eukaryotic transcription turns 50, Cell 179, 808 (2019). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
