Ethylene enhances MdMAPK3-mediated phosphorylation of MdNAC72 to promote apple fruit softening
- PMID: 37132483
- PMCID: PMC10396387
- DOI: 10.1093/plcell/koad122
Ethylene enhances MdMAPK3-mediated phosphorylation of MdNAC72 to promote apple fruit softening
Abstract
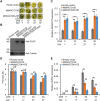
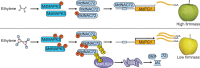
The phytohormone ethylene plays an important role in promoting the softening of climacteric fruits, such as apples (Malus domestica); however, important aspects of the underlying regulatory mechanisms are not well understood. In this study, we identified apple MITOGEN-ACTIVATED PROTEIN KINASE 3 (MdMAPK3) as an important positive regulator of ethylene-induced apple fruit softening during storage. Specifically, we show that MdMAPK3 interacts with and phosphorylates the transcription factor NAM-ATAF1/2-CUC2 72 (MdNAC72), which functions as a transcriptional repressor of the cell wall degradation-related gene POLYGALACTURONASE1 (MdPG1). The increase in MdMAPK3 kinase activity was induced by ethylene, which promoted the phosphorylation of MdNAC72 by MdMAPK3. Additionally, MdPUB24 functions as an E3 ubiquitin ligase to ubiquitinate MdNAC72, resulting in its degradation via the 26S proteasome pathway, which was enhanced by ethylene-induced phosphorylation of MdNAC72 by MdMAPK3. The degradation of MdNAC72 increased the expression of MdPG1, which in turn promoted apple fruit softening. Notably, using variants of MdNAC72 that were mutated at specific phosphorylation sites, we observed that the phosphorylation state of MdNAC72 affected apple fruit softening during storage. This study thus reveals that the ethylene-MdMAPK3-MdNAC72-MdPUB24 module is involved in ethylene-induced apple fruit softening, providing insights into climacteric fruit softening.
© The Author(s) 2023. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. The authors declare no conflict of interest.
Figures




References
-
- Atkinson RG, Sutherland PW, Johnston SL, Gunaseelan K, Hallett IC, Mitra D, Brummell DA, Schröder R, Johnston JW, Schaffer RJ. Down-regulation of POLYGALACTURONASE1 alters firmness, tensile strength and water loss in apple (Malus x domestica) fruit. BMC Plant Biol. 2012:12(1):129. 10.1186/1471-2229-12-129 - DOI - PMC - PubMed
-
- Berriri S, Garcia AV, dit Frey NF, Rozhon W, Pateyron S, Leonhardt N, Montillet J-L, Leung J, Hirt H, Colcombet J. Constitutively active mitogen-activated protein kinase versions reveal functions of Arabidopsis MPK4 in pathogen defense signaling. Plant Cell. 2012:24(10):4281–4293. 10.1105/tpc.112.101253 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

