Improved T cell receptor antigen pairing through data-driven filtering of sequencing information from single cells
- PMID: 37133356
- PMCID: PMC10156162
- DOI: 10.7554/eLife.81810
Improved T cell receptor antigen pairing through data-driven filtering of sequencing information from single cells
Abstract
Novel single-cell-based technologies hold the promise of matching T cell receptor (TCR) sequences with their cognate peptide-MHC recognition motif in a high-throughput manner. Parallel capture of TCR transcripts and peptide-MHC is enabled through the use of reagents labeled with DNA barcodes. However, analysis and annotation of such single-cell sequencing (SCseq) data are challenged by dropout, random noise, and other technical artifacts that must be carefully handled in the downstream processing steps. We here propose a rational, data-driven method termed ITRAP (improved T cell Receptor Antigen Paring) to deal with these challenges, filtering away likely artifacts, and enable the generation of large sets of TCR-pMHC sequence data with a high degree of specificity and sensitivity, thus outputting the most likely pMHC target per T cell. We have validated this approach across 10 different virus-specific T cell responses in 16 healthy donors. Across these samples, we have identified up to 1494 high-confident TCR-pMHC pairs derived from 4135 single cells.
Keywords: T cell receptor; computational biology; epitope specificity; evaluation methods; human; immunology; inflammation; sequence similarity; single-cell immune profiling; systems biology.
© 2023, Povlsen, Bentzen et al.
Conflict of interest statement
HP, MK, LJ, MN No competing interests declared, AB, SH AKB and SRH are co-inventors on a patent covering the use of DNA barcode labeled MHC multimers (WO2015185067 and WO2015188839), which is licensed to Immudex
Figures
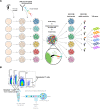
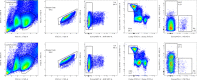

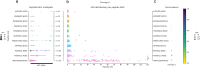

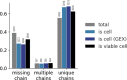




Update of
- doi: 10.1101/2022.08.31.506001
References
-
- 10xGenomics Cell Ranger Installation -Software -Single Cell Immune Profiling -Official 10x Genomics Support. 2022a. [July 12, 2022]. https://support.10xgenomics.com/single-cell-vdj/software/pipelines/lates...
-
- 10xGenomics V(D)J Cell Calling Algorithm -Software -Single Cell Immune Profiling -Official 10x Genomics Support. 2022b. [July 12, 2022]. https://support.10xgenomics.com/single-cell-vdj/software/pipelines/lates...
-
- Acha-Orbea H, Mitchell DJ, Timmermann L, Wraith DC, Tausch GS, Waldor MK, Zamvil SS, McDevitt HO, Steinman L. Limited heterogeneity of T cell receptors from lymphocytes mediating autoimmune encephalomyelitis allows specific immune intervention. Cell. 1988;54:263–273. doi: 10.1016/0092-8674(88)90558-2. - DOI - PubMed
-
- Bagaev DV, Vroomans RMA, Samir J, Stervbo U, Rius C, Dolton G, Greenshields-Watson A, Attaf M, Egorov ES, Zvyagin IV, Babel N, Cole DK, Godkin AJ, Sewell AK, Kesmir C, Chudakov DM, Luciani F, Shugay M. VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Research. 2020;48:D1057–D1062. doi: 10.1093/nar/gkz874. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

