Identification of New Compounds against PRRSV Infection by Directly Targeting CD163
- PMID: 37133376
- PMCID: PMC10231194
- DOI: 10.1128/jvi.00054-23
Identification of New Compounds against PRRSV Infection by Directly Targeting CD163
Abstract
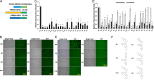
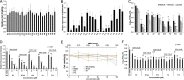
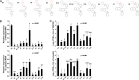
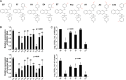
The porcine reproductive and respiratory syndrome viruses (PRRSV) led to a global panzootic and huge economical losses to the pork industry. PRRSV targets the scavenger receptor CD163 for productive infection. However, currently no effective treatment is available to control the spread of this disease. Using bimolecular fluorescence complementation (BiFC) assays, we screened a set of small molecules potentially targeting the scavenger receptor cysteine-rich domain 5 (SRCR5) of CD163. We found that the assay examining protein-protein interactions (PPI) between PRRSV glycoprotein 4 (GP4) and the CD163-SRCR5 domain mainly identifies compounds that potently inhibit PRRSV infection, while examining the PPI between PRRSV-GP2a and the SRCR5 domain maximized the identification of positive compounds, including additional ones with various antiviral capabilities. These positive compounds significantly inhibited both types 1 and 2 PRRSV infection of porcine alveolar macrophages. We confirmed that the highly active compounds physically bind to the CD163-SRCR5 protein, with dissociation constant (KD) values ranging from 28 to 39 μM. Structure-activity-relationship (SAR) analysis revealed that although both the 3-(morpholinosulfonyl)anilino and benzenesulfonamide moieties in these compounds are critical for the potency to inhibit PRRSV infection, the morpholinosulfonyl group can be replaced by chlorine substituents without significant loss of antiviral potency. Our study established a system for throughput screening of natural or synthetic compounds highly effective on blocking of PRRSV infection and shed light on further SAR modification of these compounds. IMPORTANCE Porcine reproductive and respiratory syndrome virus (PRRSV) causes significant economic losses to the swine industry worldwide. Current vaccines cannot provide cross protection against different strains, and there are no effective treatments available to hamper the spread of this disease. In this study, we identified a group of new small molecules that can inhibit the PRRSV interaction with its specific receptor CD163 and dramatically block the infection of both types 1 and type 2 PRRSVs to host cells. We also demonstrated the physical association of these compounds with the SRCR5 domain of CD163. In addition, molecular docking and structure-activity relationship analyses provided new insights for the CD163/PRRSV glycoprotein interaction and further improvement of these compounds against PRRSV infection.
Keywords: CD163; PRRSV; SRCR5; antiviral treatment; small molecule compound.
Conflict of interest statement
A provisional patent application was filed by the University of Connecticut and Atomwise, Inc.
Figures

Similar articles
-
Small molecules block the interaction between porcine reproductive and respiratory syndrome virus and CD163 receptor and the infection of pig cells.Virol J. 2020 Jul 30;17(1):116. doi: 10.1186/s12985-020-01361-7. Virol J. 2020. PMID: 32727587 Free PMC article.
-
Pigs Lacking the Scavenger Receptor Cysteine-Rich Domain 5 of CD163 Are Resistant to Porcine Reproductive and Respiratory Syndrome Virus 1 Infection.J Virol. 2018 Jul 31;92(16):e00415-18. doi: 10.1128/JVI.00415-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29925651 Free PMC article.
-
The Crystal Structure of the Fifth Scavenger Receptor Cysteine-Rich Domain of Porcine CD163 Reveals an Important Residue Involved in Porcine Reproductive and Respiratory Syndrome Virus Infection.J Virol. 2017 Jan 18;91(3):e01897-16. doi: 10.1128/JVI.01897-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27881657 Free PMC article.
-
Role of CD163 in PRRSV infection.Virology. 2024 Dec;600:110262. doi: 10.1016/j.virol.2024.110262. Epub 2024 Oct 16. Virology. 2024. PMID: 39423600 Review.
-
PRRS virus receptors and their role for pathogenesis.Vet Microbiol. 2015 Jun 12;177(3-4):229-41. doi: 10.1016/j.vetmic.2015.04.002. Epub 2015 Apr 13. Vet Microbiol. 2015. PMID: 25912022 Review.
Cited by
-
A chimeric strain of porcine reproductive and respiratory syndrome virus 2 derived from HP-PRRSV and NADC30-like PRRSV confers cross-protection against both strains.Vet Res. 2024 Oct 7;55(1):132. doi: 10.1186/s13567-024-01390-y. Vet Res. 2024. PMID: 39375803 Free PMC article.
-
Monoclonal Antibodies Targeting Porcine Macrophages Are Able to Inhibit the Cell Entry of Macrophage-Tropic Viruses (PRRSV and ASFV).Viruses. 2025 Jan 24;17(2):167. doi: 10.3390/v17020167. Viruses. 2025. PMID: 40006922 Free PMC article.
-
Comprehensive Review on Bimolecular Fluorescence Complementation and Its Application in Deciphering Protein-Protein Interactions in Cell Signaling Pathways.Biomolecules. 2024 Jul 17;14(7):859. doi: 10.3390/biom14070859. Biomolecules. 2024. PMID: 39062573 Free PMC article. Review.
-
Formononetin and mizoribine inhibit Porcine Reproductive and Respiratory Syndrome Virus replication in vitro.Front Nutr. 2025 Mar 24;11:1501685. doi: 10.3389/fnut.2024.1501685. eCollection 2024. Front Nutr. 2025. PMID: 40196742 Free PMC article.
-
Analysis of whole transcriptome reveals the immune response to porcine reproductive and respiratory syndrome virus infection and tylvalosin tartrate treatment in the porcine alveolar macrophages.Front Immunol. 2025 Jan 13;15:1506371. doi: 10.3389/fimmu.2024.1506371. eCollection 2024. Front Immunol. 2025. PMID: 39872536 Free PMC article.
References
-
- Pork-Checkoff. 2012. Checkoff research helps wage war on PRRS. Pork Checkoff Special Edition Report 8. https://www.nationalhogfarmer.com/health/checkoff-research-helps-wage-wa....
-
- Paz XD. 17 August 2015. PRRS cost for the European swine industry. Pig333, Ripoll, Spain. https://www.pig333.com/articles/prrs-cost-for-the-european-swine-industr....
-
- Halbur PG. 1998. “PRRS Plus”–PRRS virus infection in combination with other agents. Hypothesis 1997:1998b–2000.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

