CkP1 bacteriophage, a S16-like myovirus that recognizes Citrobacter koseri lipopolysaccharide through its long tail fibers
- PMID: 37133800
- PMCID: PMC10175313
- DOI: 10.1007/s00253-023-12547-8
CkP1 bacteriophage, a S16-like myovirus that recognizes Citrobacter koseri lipopolysaccharide through its long tail fibers
Abstract
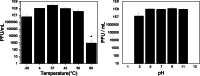
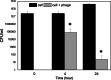
Citrobacter koseri is an emerging Gram-negative bacterial pathogen, which causes urinary tract infections. We isolated and characterized a novel S16-like myovirus CKP1 (vB_CkoM_CkP1), infecting C. koseri. CkP1 has a host range covering the whole C. koseri species, i.e., all strains that were tested, but does not infect other species. Its linear 168,463-bp genome contains 291 coding sequences, sharing sequence similarity with the Salmonella phage S16. Based on surface plasmon resonance and recombinant green florescence protein fusions, the tail fiber (gp267) was shown to decorate C. koseri cells, binding with a nanomolar affinity, without the need of accessory proteins. Both phage and the tail fiber specifically bind to bacterial cells by the lipopolysaccharide polymer. We further demonstrate that CkP1 is highly stable towards different environmental conditions of pH and temperatures and is able to control C. koseri cells in urine samples. Altogether, CkP1 features optimal in vitro characteristics to be used both as a control and detection agent towards drug-resistant C. koseri infections. KEY POINTS: • CkP1 infects all C. koseri strains tested • CkP1 recognizes C. koseri lipopolysaccharide through its long tail fiber • Both phage CkP1 and its tail fiber can be used to treat or detect C. koseri pathogens.
Keywords: Bacterial infection; Bacteriophage; Citrobacter; Control; Diagnostics; Long tail fiber.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Characterization and genome sequencing of a Citrobacter freundii phage CfP1 harboring a lysin active against multidrug-resistant isolates.Appl Microbiol Biotechnol. 2016 Dec;100(24):10543-10553. doi: 10.1007/s00253-016-7858-0. Epub 2016 Sep 28. Appl Microbiol Biotechnol. 2016. PMID: 27683211
-
Long tail fibres of the novel broad-host-range T-even bacteriophage S16 specifically recognize Salmonella OmpC.Mol Microbiol. 2013 Feb;87(4):818-34. doi: 10.1111/mmi.12134. Epub 2013 Jan 15. Mol Microbiol. 2013. PMID: 23289425
-
Modified Bacteriophage Tail Fiber Proteins for Labeling, Immobilization, Capture, and Detection of Bacteria.Methods Mol Biol. 2019;1918:67-86. doi: 10.1007/978-1-4939-9000-9_6. Methods Mol Biol. 2019. PMID: 30580400
-
Optimum management of Citrobacter koseri infection.Expert Rev Anti Infect Ther. 2014 Sep;12(9):1137-42. doi: 10.1586/14787210.2014.944505. Epub 2014 Aug 2. Expert Rev Anti Infect Ther. 2014. PMID: 25088467 Review.
-
Understanding Bacteriophage Tail Fiber Interaction with Host Surface Receptor: The Key "Blueprint" for Reprogramming Phage Host Range.Int J Mol Sci. 2022 Oct 12;23(20):12146. doi: 10.3390/ijms232012146. Int J Mol Sci. 2022. PMID: 36292999 Free PMC article. Review.
References
-
- Ackermann HW. Frequency of morphological phage descriptions in 1995. Adv Virol. 1996;141(2):209–218. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. - PubMed
-
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. - PMC - PubMed
-
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

