Acetylcarnitine shuttling links mitochondrial metabolism to histone acetylation and lipogenesis
- PMID: 37134161
- PMCID: PMC10156126
- DOI: 10.1126/sciadv.adf0115
Acetylcarnitine shuttling links mitochondrial metabolism to histone acetylation and lipogenesis
Abstract
The metabolite acetyl-CoA is necessary for both lipid synthesis in the cytosol and histone acetylation in the nucleus. The two canonical precursors to acetyl-CoA in the nuclear-cytoplasmic compartment are citrate and acetate, which are processed to acetyl-CoA by ATP-citrate lyase (ACLY) and acyl-CoA synthetase short-chain 2 (ACSS2), respectively. It is unclear whether other substantial routes to nuclear-cytosolic acetyl-CoA exist. To investigate this, we generated cancer cell lines lacking both ACLY and ACSS2 [double knockout (DKO) cells]. Using stable isotope tracing, we show that both glucose and fatty acids contribute to acetyl-CoA pools and histone acetylation in DKO cells and that acetylcarnitine shuttling can transfer two-carbon units from mitochondria to cytosol. Further, in the absence of ACLY, glucose can feed fatty acid synthesis in a carnitine responsive and carnitine acetyltransferase (CrAT)-dependent manner. The data define acetylcarnitine as an ACLY- and ACSS2-independent precursor to nuclear-cytosolic acetyl-CoA that can support acetylation, fatty acid synthesis, and cell growth.
Figures
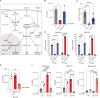
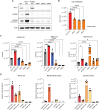





References
-
- S. Zhao, A. M. Torres, R. A. Henry, S. Trefely, M. Wallace, J. V. Lee, A. Carrer, A. Sengupta, S. L. Campbell, Y.-M. Kuo, A. J. Frey, N. Meurs, J. M. Viola, I. A. Blair, A. M. Weljie, C. M. Metallo, N. W. Snyder, A. J. Andrews, K. E. Wellen, ATP-citrate lyase controls a glucose-to-acetate metabolic switch. Cell Rep. 17, 1037–1052 (2016). - PMC - PubMed
MeSH terms
Substances
Grants and funding
- P30 AR069589/AR/NIAMS NIH HHS/United States
- K00 CA212455/CA/NCI NIH HHS/United States
- T32 GM142606/GM/NIGMS NIH HHS/United States
- R01 CA262055/CA/NCI NIH HHS/United States
- R01 GM132261/GM/NIGMS NIH HHS/United States
- R01 CA174761/CA/NCI NIH HHS/United States
- P30 ES013508/ES/NIEHS NIH HHS/United States
- T32 AR007465/AR/NIAMS NIH HHS/United States
- T32 CA115299/CA/NCI NIH HHS/United States
- T32 GM007229/GM/NIGMS NIH HHS/United States
- R01 DK116005/DK/NIDDK NIH HHS/United States
- K12 GM081259/GM/NIGMS NIH HHS/United States
- R01 CA248315/CA/NCI NIH HHS/United States
- R01 CA228339/CA/NCI NIH HHS/United States
- R01 AR077615/AR/NIAMS NIH HHS/United States
- R01 CA259111/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

